Difference between revisions of "Part:BBa K729003"
Sednanalien (Talk | contribs) (→Result) |
Sednanalien (Talk | contribs) (→Congo Red Agar Assay) |
||
| Line 38: | Line 38: | ||
! Control (W3110) !! BBa_K729017 | ! Control (W3110) !! BBa_K729017 | ||
|- | |- | ||
| − | | [[ | + | | [[Image:UniversityCollegeLondon_Curli_Congo_ Red_Control.jpg|475px]] || [[Image:UniversityCollegeLondon_Curli_Congo_ Red_K729017.jpg|475px]] |
|} | |} | ||
Revision as of 10:34, 30 September 2012
csgBAEFG operon
Stand alone csgBAEFG operon for curli synthesis. Adapted from Lyon 2011. User experience uploaded on the original page here: https://parts.igem.org/Part:BBa_K540000:Experience#User_Reviews.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1683
- 1000COMPATIBLE WITH RFC[1000]
Characterisation
The characterisation of curli expression will be done via a Congo Red agar assay. Congo Red is a diazo dye that causes cells expressing curlis to be stained red. Hence, Congo Red agar provides a convenient assay to determine if the expression of curlis has been successful.
The linear structure of the amyloid curli fibril allows the formation of hydrogen bonds with the Congo Red agar, while alternative structures are forced to form ionic bonds. These ionic bonds allow the rapid dissipation of the dye colour, resulting in the formation of pale plaques where curli negative growth occurs. The hydrogen bonds do not facilitate this removal of the dye colour, hence their growth as red colonies. This assay acts as a highly effective indicator of Curli expression.
Besides detecting the expression of curlis, we also want to ascertain the capabilities of the biofilm formed by the curliated cells and more specifically the strength of their adhesion to the plastic itself.
As such, we have worked with the Engineering Faculty Rapid Design and Fabrication Facility (RDFF) to design and construct a small scale shear device which is used to quantitatively analyse the critical shear force that can be applied to biofilms expressing the UCL '12 curli construct (BBa_K729018) before their dettachment from the surface of plastics, and as such tell us how strong the bond between cell and plastic are due to the curli fibrils.
The approach taken in generating and using this device as a characteristion method is truly novel. Where previously characterisation for a construct such as this has been very qualitative, merely detecting the presence of cells or biofilm or at most measuring biofilm thickness, this new apparatus creates a variable shear gradient across the surface of the biofilm at a single rotational speed. This means that the interface between where biofilm is present and where it is not after the test is used to calculate (using the equation below) and quantify the binding strength of the surface proteins.
Variations
Two additional variations on the original biobrick were designed. These include a T7 and constitutive promoter.
Result
Congo Red Agar Assay
We have used a congo red agar assay to determine if our curli BioBrick transformed cells have successfully expressed the amyloid fibrins. Congo red is a diazo dye that binds to the curli fibrins, thereby causing cells positive for curlis to appear red when grown on this agar.
As can be seen from plates below, our cells transformed with BBa_K729017 display a red tinge to their colonies, while the untransformed W3110 cells do not. We hence conclude that we have successfully transformed our cells to produce curlis.
| Control (W3110) | BBa_K729017 |
|---|---|
 |

|
Shear Force Assay
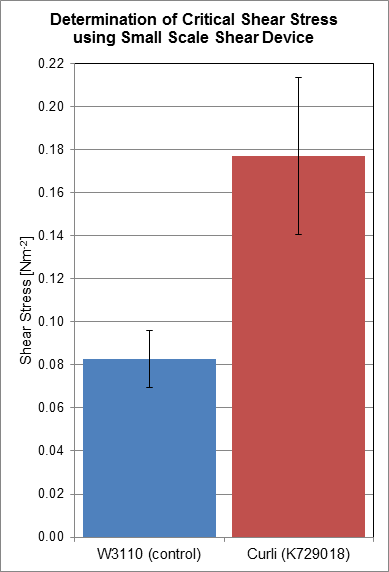
Using a rotary shear device, we have obtained data relating to the shear resistance of our curliated biofilms. Over a 24 hour period, biofilms were allowed to grow on the surface of plastic discs, which were then subjected to shear forces in the rotary shear device. By finding the critical shear radius, the point at which cells start to detach from the plastic discs, we have calculated the critical shear stresses that our biofilms are capable of resisting.
Based on the results we have obtained, we have determined that the biofilms produced by cells transformed with BBa_K729018 have a two-fold shear resistance when compared to the biofilms of the control cell line W3110. This gives corroborating evidence that our curli BioBrick is functioning as intended, as curli generated biofilms are more resistant to shear forces, and detach less readily.