Difference between revisions of "Part:BBa K633015"
Faustortiz (Talk | contribs) |
|||
| Line 2: | Line 2: | ||
<partinfo>BBa_K633015 short</partinfo> | <partinfo>BBa_K633015 short</partinfo> | ||
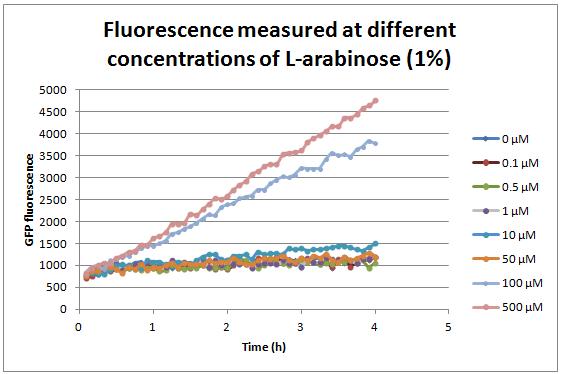
| − | + | In order to find suitable arabinose concentrations to induce expression of the construct, the protein coding sequences were substituted by a GFP reporter, BBa_e0040. A double terminator, BBa_e0014, was added as well. The expression vehicle was Escherichia coli, BW27783 strain, due to its inability to metabolize arabinose. Fluorescence was measured in transformed E. coli cultures induced with different concentrations of L-arabinose (1%), following the protocol used by Cambridge (2009) and Tec-Monterrey (2010). | |
| − | + | [[Image:Induction_gfp.jpg]] | |
| − | + | ||
| − | + | The chart shows that at low concentrations of arabinose, poor induction levels are obtained since fluorescence changes very little over time. A noticeable increase of fluorescence is viewed from a concentration of 100 µM onwards. Similar results were found in BBa_I0500 characterization by Cambridge (2011) and Groningen (2011), proving further the effectiveness of using this concentration of arabinose as a minimum to induce expression of the construct. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 14:54, 17 October 2011
AraC+Pbad promoter+RBS+ Ompa&lpp signal peptide+Sacc
In order to find suitable arabinose concentrations to induce expression of the construct, the protein coding sequences were substituted by a GFP reporter, BBa_e0040. A double terminator, BBa_e0014, was added as well. The expression vehicle was Escherichia coli, BW27783 strain, due to its inability to metabolize arabinose. Fluorescence was measured in transformed E. coli cultures induced with different concentrations of L-arabinose (1%), following the protocol used by Cambridge (2009) and Tec-Monterrey (2010).
The chart shows that at low concentrations of arabinose, poor induction levels are obtained since fluorescence changes very little over time. A noticeable increase of fluorescence is viewed from a concentration of 100 µM onwards. Similar results were found in BBa_I0500 characterization by Cambridge (2011) and Groningen (2011), proving further the effectiveness of using this concentration of arabinose as a minimum to induce expression of the construct.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1247
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1187
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
Illegal AgeI site found at 2657 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2357
Illegal SapI site found at 961