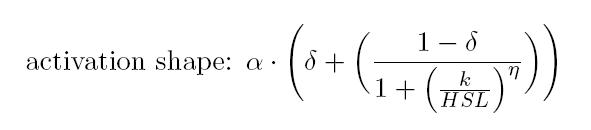Difference between revisions of "Part:BBa T9002:Experience"
Elena moreno (Talk | contribs) |
(Reformatting to separate reviews) |
||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | + | This experience page is provided so that any user may enter their |
| + | |||
| + | experience using this part.<BR>Please enter | ||
how you used this part and how it worked out. | how you used this part and how it worked out. | ||
| − | + | =Applications of BBa_T9002= | |
| + | |||
| + | =User Reviews= | ||
| + | |||
| + | === UNIPV-Pavia iGEM 2011 === | ||
| − | |||
| − | |||
| − | |||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
| − | |width=' | + | |width='20%' valign='top'| |
| − | <partinfo>BBa_T9002 AddReview | + | <partinfo>BBa_T9002 AddReview 5</partinfo> |
| − | <I> | + | <I>UNIPV-Pavia iGEM 2011</I> |
| − | |width=' | + | |width='50%' valign='top'| |
| − | + | ||
| − | + | ''Antiquity. This review comes from the old result system and indicates that this part worked in some test.'' | |
| − | + | ||
'''2009 DNA Distribution quality control''' | '''2009 DNA Distribution quality control''' | ||
| − | The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was completely confirmed, while iGEM QC results classified it as "inconsistent". DNA was resuspended from well 9A, kit plate 2, transformed in TOP10 E. coli and amplified inoculating a single colony from the grown LB agar plate in LB medium. Finally DNA has been miniprepped from the grown culture and sent to a BMR Genomics (Padova, Italy) for sequencing. | + | The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was |
| + | |||
| + | completely confirmed, while iGEM QC results classified it as | ||
| + | |||
| + | "inconsistent". DNA was resuspended from well 9A, kit plate 2, | ||
| + | |||
| + | transformed in TOP10 E. coli and amplified inoculating a single colony | ||
| + | |||
| + | from the grown LB agar plate in LB medium. Finally DNA has been | ||
| + | |||
| + | miniprepped from the grown culture and sent to a BMR Genomics (Padova, | ||
| + | |||
| + | Italy) for sequencing. | ||
'''Experimental measurements''' | '''Experimental measurements''' | ||
| − | The UNIPV-Pavia iGEM team tested T9002 BioBrick in several working | + | The UNIPV-Pavia iGEM team tested T9002 BioBrick in several working |
| − | The Brown iGEM team conducted tests on this part in the summer of 2007. The results are depicted in the graphs below. | + | conditions. Results are reported in <partinfo>BBa_F2620</partinfo> |
| + | |||
| + | Experience page. | ||
| + | |||
| + | The Brown iGEM team conducted tests on this part in the summer of 2007. | ||
| + | |||
| + | The results are depicted in the graphs below. | ||
[[Image:AHL.JPG|700px]] | [[Image:AHL.JPG|700px]] | ||
| − | The first graph indicates that until a critical point is reached, | + | The first graph indicates that until a critical point is reached, |
| + | increasing AHL concentration does increase GFP output. This is most | ||
| − | + | easily noticeable between the concentrations of 20 nM, after which | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | point the amount of GFP produced by cells begins to decrease. This may | |
| + | |||
| + | be due to one of two things: AHL quenches the signal from GFP, or too | ||
| + | |||
| + | much AHL disrupts the cell's functions in a way that either kills it or | ||
| + | |||
| + | prevents it from making as much GFP. This second hypothesis is | ||
| + | |||
| + | partially confirmed by the second graph, which shows that adding more | ||
| + | |||
| + | than 20 nM AHL causes a decline in cell density. On each graph, the | ||
| + | |||
| + | different colored lines represent different time points after AHL was | ||
| + | |||
| + | added to the cells. They are 4 hours, 5 hours, etc. | ||
| + | |||
| + | ---- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<html> | <html> | ||
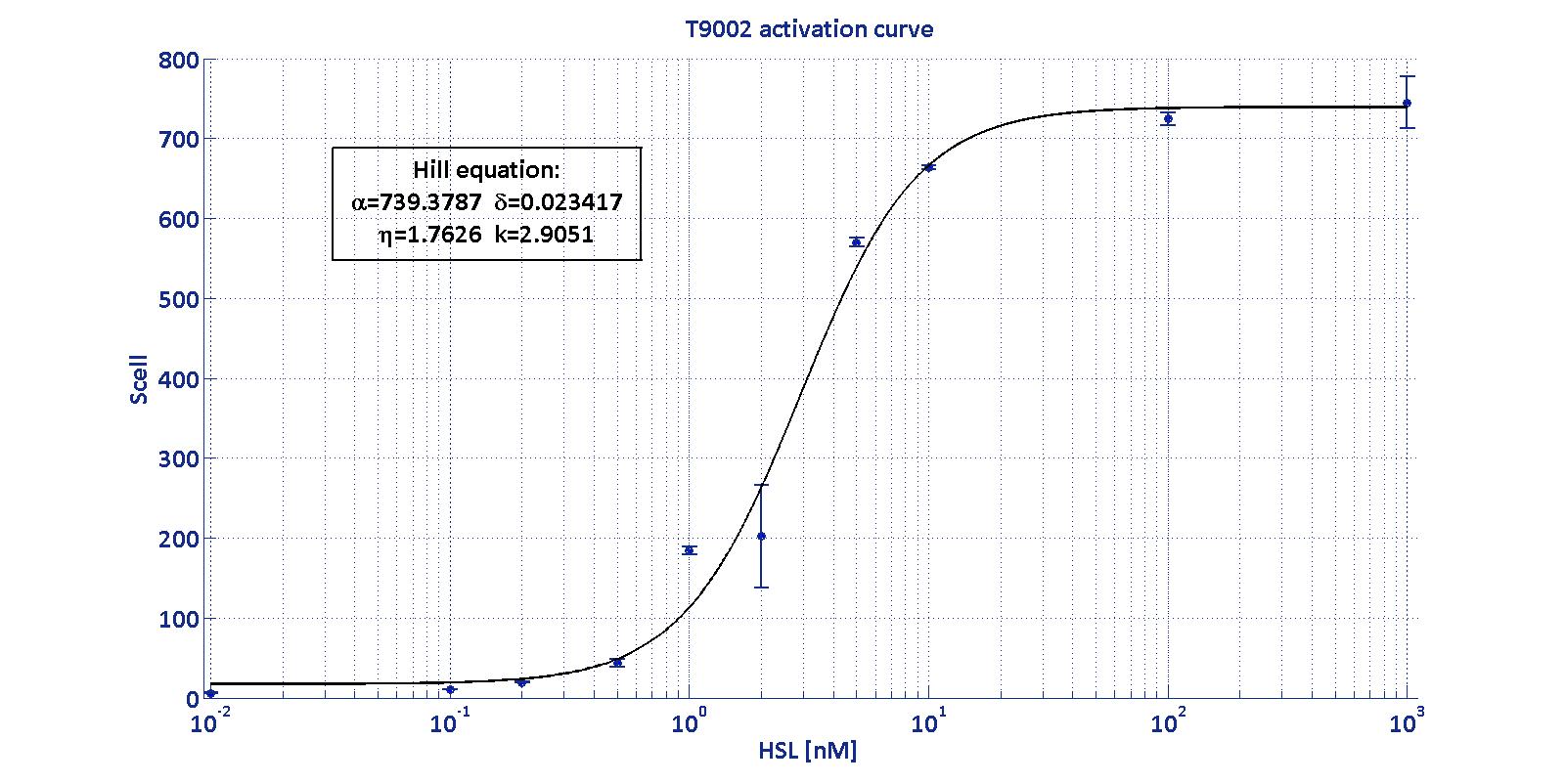
| − | BioBrick <a href="https://parts.igem.org/Part:BBa_T9002">BBa_T9002</a> is an HSL biosensor, which provides a non linear relationship between HSL input and S<sub>cell</sub> output. More precisely, the characteristic sigmoidal curve requires synthetic parameters for its accurate identification. These are the minimum and maximum values, the swtich point (i.e., the curve inflection point), and the upper and lower boundaries of linearity. This biosensor revealed greatly reliable, providing measurement repeatability and minimal experimental noise. Referring to its activation formula, the calibration curve is shown below.<br></div> | + | BioBrick <a href="https://parts.igem.org/Part:BBa_T9002">BBa_T9002</a> |
| + | |||
| + | is an HSL biosensor, which provides a non linear relationship between | ||
| + | |||
| + | HSL input and S<sub>cell</sub> output. More precisely, the | ||
| + | |||
| + | characteristic sigmoidal curve requires synthetic parameters for its | ||
| + | |||
| + | accurate identification. These are the minimum and maximum values, the | ||
| + | |||
| + | swtich point (i.e., the curve inflection point), and the upper and | ||
| + | |||
| + | lower boundaries of linearity. This biosensor revealed greatly | ||
| + | |||
| + | reliable, providing measurement repeatability and minimal experimental | ||
| + | |||
| + | noise. Referring to its activation formula, the calibration curve is | ||
| + | |||
| + | shown below.<br></div> | ||
<br> | <br> | ||
<div style='text-align:center'> | <div style='text-align:center'> | ||
| − | <div class="thumbinner" style="width:100%;"> <a href="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg"><img alt="" src="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg" class="thumbimage" width="47%" height="50%"></a></div> | + | <div class="thumbinner" style="width:100%;"> <a |
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg"><img | ||
| + | |||
| + | alt="" src="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg" | ||
| + | |||
| + | class="thumbimage" width="47%" height="50%"></a></div> | ||
</div> | </div> | ||
<br> | <br> | ||
<div style='text-align:center'> | <div style='text-align:center'> | ||
| − | <div class="thumbinner" style="width:100%;"> <a href="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg"><img alt="" src="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg" class="thumbimage" width="100%"></a></div> | + | <div class="thumbinner" style="width:100%;"> <a |
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg"><img | ||
| + | |||
| + | alt="" src="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg" | ||
| + | |||
| + | class="thumbimage" width="100%"></a></div> | ||
</div> | </div> | ||
<center> | <center> | ||
| Line 94: | Line 136: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | <div align="justify">In order to determine the threshold sensitivity of T9002 biosensor, experiments were performed with several HSL inductions minimally interspaced in the region of low detectability. Hypothesizing that the inducer is 1:20 diluted (as for all of our tests), the minimum detectable HSL concentration is 3 nM.</div> | + | <div align="justify">In order to determine the threshold sensitivity of |
| + | |||
| + | T9002 biosensor, experiments were performed with several HSL inductions | ||
| + | |||
| + | minimally interspaced in the region of low detectability. Hypothesizing | ||
| + | |||
| + | that the inducer is 1:20 diluted (as for all of our tests), the minimum | ||
| + | |||
| + | detectable HSL concentration is 3 nM.</div> | ||
<br><br> | <br><br> | ||
<a name='t9002PV11'></a> | <a name='t9002PV11'></a> | ||
| − | This biosensor was used to measure HSL concentration for parts | + | This biosensor was used to measure HSL concentration for parts |
| − | <div class="center"><div class="thumbinner" style="width: 650px;"><a href="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG"class=" | + | producing or degrading this signalling molecule. For each of these |
| + | |||
| + | experiments, a calibration curve of the biosensor was built, inducing | ||
| + | |||
| + | it with known HSL concentrations, evaluating for each of them the | ||
| + | |||
| + | S<sub>cell</sub> signal and finally estimating the Hill curve | ||
| + | |||
| + | parameters. Once identified the parameters, the unknown concentration | ||
| + | |||
| + | of a sample can be evaluated from its S<sub>cell</sub> (provided that | ||
| + | |||
| + | it has a value included in the linear zone of the biosensor), as shown | ||
| + | |||
| + | below: | ||
| + | |||
| + | <div class="center"><div class="thumbinner" style="width: 650px;"><a | ||
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG" | ||
| + | |||
| + | class="image"><img alt="" | ||
| + | |||
| + | src="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG"class="thumbim | ||
| + | |||
| + | age" width="40%"></a></div></div> | ||
<br> | <br> | ||
| − | It is necessary, then, to multiply the measurement for the dilution factor used (in our experiments it was 20). | + | It is necessary, then, to multiply the measurement for the dilution |
| + | |||
| + | factor used (in our experiments it was 20). | ||
</html> | </html> | ||
|} | |} | ||
| + | === Sevilla 2011 === | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='20%' valign='top'| | ||
<partinfo>BBa_T9002 AddReview 2</partinfo> | <partinfo>BBa_T9002 AddReview 2</partinfo> | ||
| − | |||
<I>Sevilla 2011</I> | <I>Sevilla 2011</I> | ||
| + | |width='50%' valign='top'| | ||
| + | |||
| + | <html><a name='t9002PV11'></a></html> | ||
| + | |||
| + | We used this construction as a model for a GFP-based characterization | ||
| + | |||
| + | method. We tried different concentrations, that turned to be too high, | ||
| + | |||
| + | for it seems that the media was quite saturated. | ||
| + | |||
| + | <nowiki>Fluorescence | ||
| − | + | ---------------------------------------------------------- O.D. | |
| − | + | (600nm)</nowiki> | |
[[Image:Fluabs.png]] | [[Image:Fluabs.png]] | ||
| Line 134: | Line 225: | ||
<!-- DON'T DELETE --><partinfo>BBa_T9002 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_T9002 EndReviews</partinfo> | ||
| + | |||
| + | |} | ||
Revision as of 17:02, 26 October 2014
This experience page is provided so that any user may enter their
experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_T9002
User Reviews
UNIPV-Pavia iGEM 2011
|
•••••
UNIPV-Pavia iGEM 2011 |
Antiquity. This review comes from the old result system and indicates that this part worked in some test.
The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was completely confirmed, while iGEM QC results classified it as "inconsistent". DNA was resuspended from well 9A, kit plate 2, transformed in TOP10 E. coli and amplified inoculating a single colony from the grown LB agar plate in LB medium. Finally DNA has been miniprepped from the grown culture and sent to a BMR Genomics (Padova, Italy) for sequencing. Experimental measurements The UNIPV-Pavia iGEM team tested T9002 BioBrick in several working conditions. Results are reported in BBa_F2620 Experience page. The Brown iGEM team conducted tests on this part in the summer of 2007. The results are depicted in the graphs below. The first graph indicates that until a critical point is reached, increasing AHL concentration does increase GFP output. This is most easily noticeable between the concentrations of 20 nM, after which point the amount of GFP produced by cells begins to decrease. This may be due to one of two things: AHL quenches the signal from GFP, or too much AHL disrupts the cell's functions in a way that either kills it or prevents it from making as much GFP. This second hypothesis is partially confirmed by the second graph, which shows that adding more than 20 nM AHL causes a decline in cell density. On each graph, the different colored lines represent different time points after AHL was added to the cells. They are 4 hours, 5 hours, etc.
BioBrick BBa_T9002
is an HSL biosensor, which provides a non linear relationship between
HSL input and Scell output. More precisely, the
characteristic sigmoidal curve requires synthetic parameters for its
accurate identification. These are the minimum and maximum values, the
swtich point (i.e., the curve inflection point), and the upper and
lower boundaries of linearity. This biosensor revealed greatly
reliable, providing measurement repeatability and minimal experimental
noise. Referring to its activation formula, the calibration curve is
shown below.
In order to determine the threshold sensitivity of
T9002 biosensor, experiments were performed with several HSL inductions
minimally interspaced in the region of low detectability. Hypothesizing
that the inducer is 1:20 diluted (as for all of our tests), the minimum
detectable HSL concentration is 3 nM.
This biosensor was used to measure HSL concentration for parts producing or degrading this signalling molecule. For each of these experiments, a calibration curve of the biosensor was built, inducing it with known HSL concentrations, evaluating for each of them the Scell signal and finally estimating the Hill curve parameters. Once identified the parameters, the unknown concentration of a sample can be evaluated from its Scell (provided that it has a value included in the linear zone of the biosensor), as shown below: It is necessary, then, to multiply the measurement for the dilution factor used (in our experiments it was 20). |
Sevilla 2011
|
••
Sevilla 2011 |
We used this construction as a model for a GFP-based characterization method. We tried different concentrations, that turned to be too high, for it seems that the media was quite saturated. Fluorescence ---------------------------------------------------------- O.D. (600nm)
UNIQ3d54f34c3ff6ac32-partinfo-00000006-QINU |