Difference between revisions of "Part:BBa K525517"
(→Usage and Biology) |
|||
| Line 59: | Line 59: | ||
<partinfo>BBa_K525517 parameters</partinfo> | <partinfo>BBa_K525517 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | <biblio> | ||
| + | #Sasaki pmid=18492046 | ||
| + | #Sasaki05a pmid=16332782 | ||
| + | </biblio> | ||
Revision as of 13:37, 17 September 2011
Fusion protein of BisdA and BisdB behind constitutive promoter
Fusion protein of BisdA and BisdB behind constitutive promoter.
Usage and Biology
Expressing this BioBrick in E. coli enables the bacterium to degrade the endocrine disruptor bisphenol A (BPA). BPA is mainly hydroxylated into the products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol. In S. bisphenolicum AO1, a total of three genes are responsible for this BPA hydroxylation: a cytochrome P450 (CYP, bisdB), a ferredoxin (Fd, bisdA) and a ferredoxin-NAD+ oxidoreductase (FNR) Sasaki05a. The three gene products act together to reduce BPA while oxidizing NADH + H+. The cytochrome P450 (BisdB) reduces the BPA and is oxidized during this reaction. BisdB in its oxidized status is reduced by the ferredoxin (BisdA) so it can reduce BPA again. The oxidized BisdA is reduced by a ferredoxin-NAD+ oxidoreductase consuming NADH + H+ so the BPA degradation can continue Sasaki05a. This electron transport chain between the three enzymes involved in BPA degradation and the BioBricks needed to enable this reaction in vivo and in vitro are shown in the following figure (please have some patience, it's an animated .gif file):
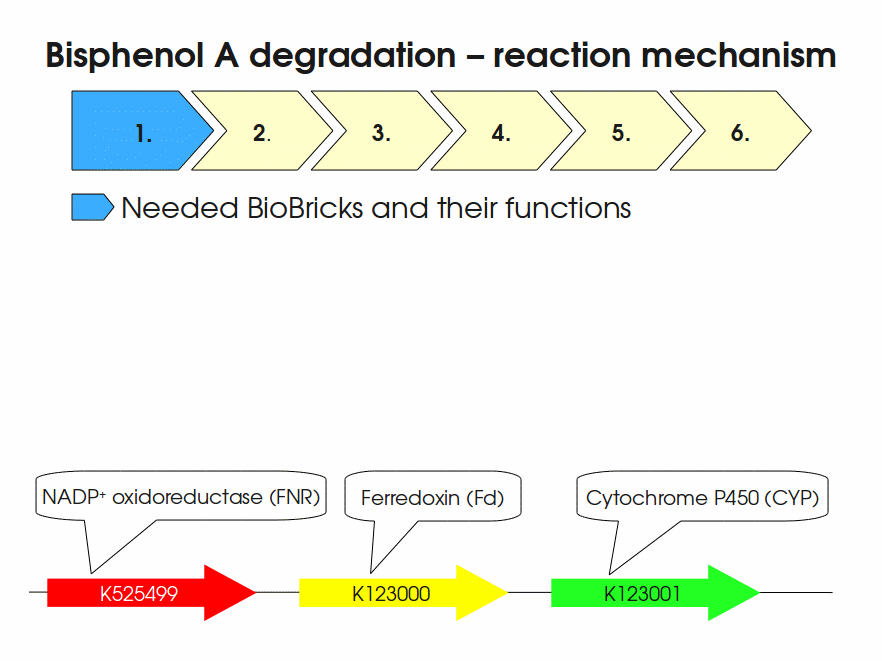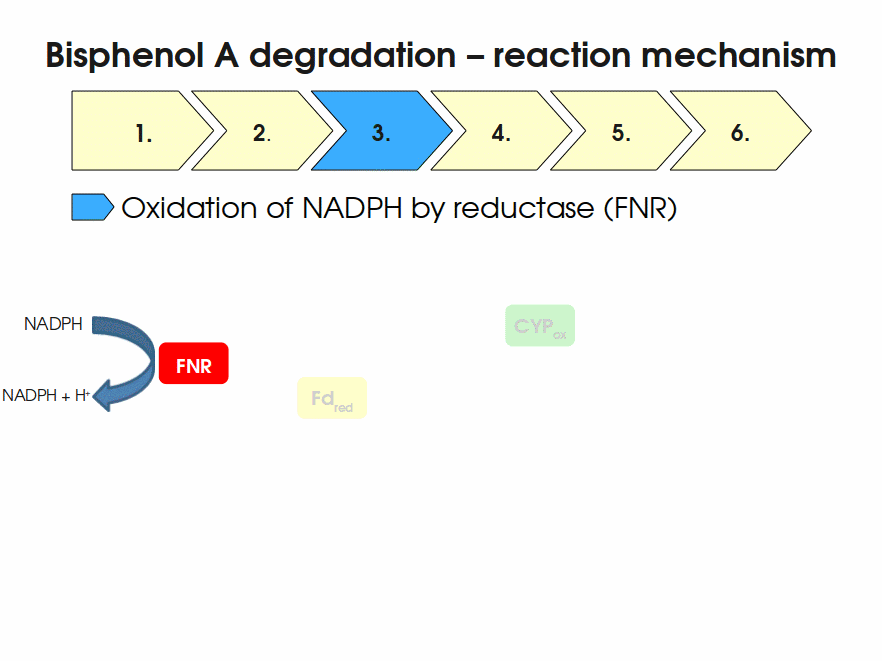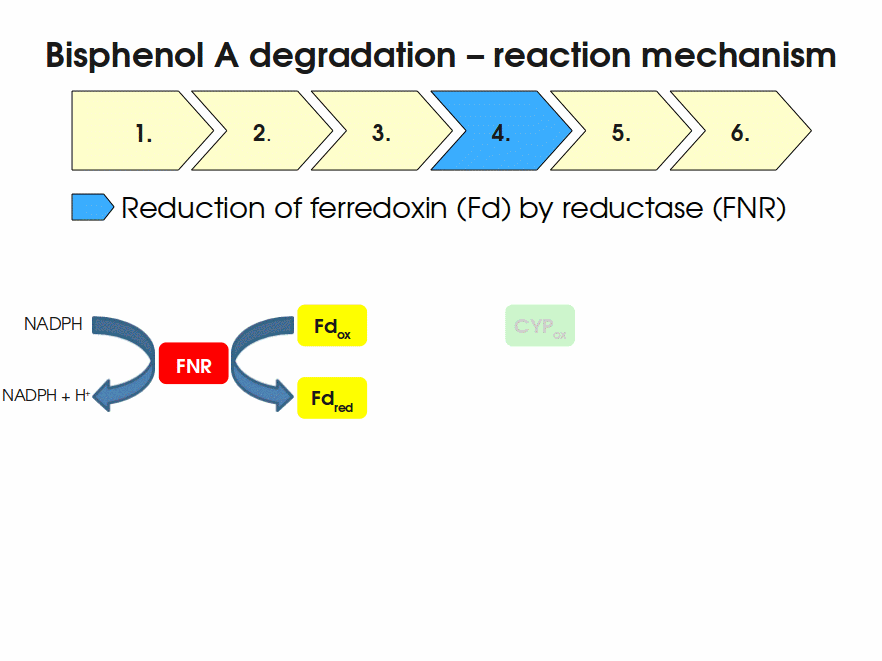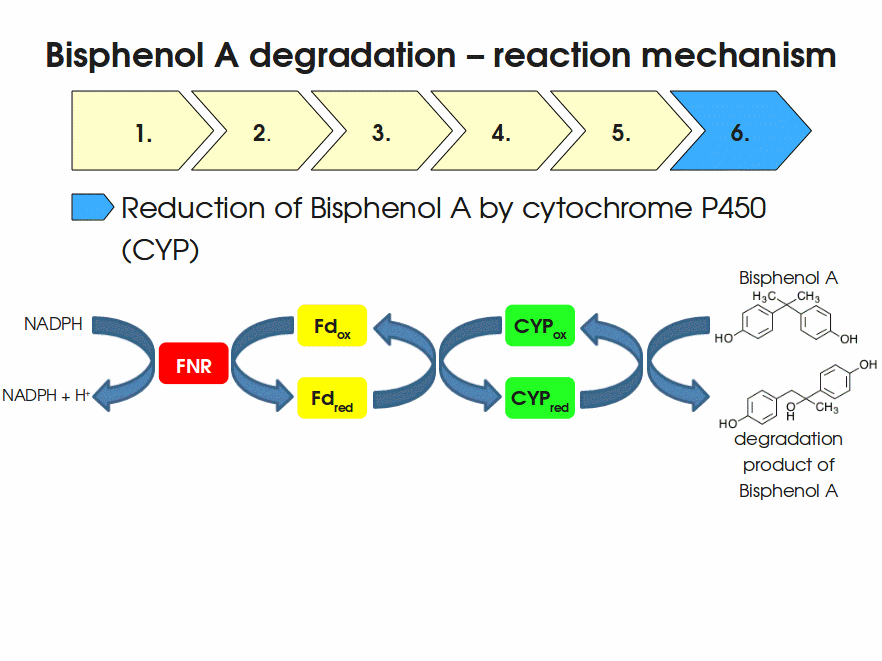
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression in E. coli | Compatibility | E. coli KRX, TOP10, MACH1, BL21(DE3) |
| Expression | Constitutive | |
| Optimal temperature | 30 °C | |
| Working BPA concentration | 120 mg L-1 (0.53 mM) | |
| Purification | Molecular weight | 59.3 kDa |
| Theoretical pI | 4.99 | |
| High absorbation | 450 nm | |
| Degradation of BPA | Completely degradation of 0.53 mM BPA | 21 - 24 h |
| Specific BPA degradation rate | 1.29 10-10 mM cell-1 |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 664
Illegal BamHI site found at 1402 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 65
Illegal AgeI site found at 1655 - 1000COMPATIBLE WITH RFC[1000]
References
<biblio>
- Sasaki pmid=18492046
- Sasaki05a pmid=16332782
</biblio>
