Difference between revisions of "Part:BBa K404164"
| (3 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K404164 short</partinfo> | <partinfo>BBa_K404164 short</partinfo> | ||
{| style="color:black" cellpadding="6" cellspacing="1" border="2" align="left" | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="left" | ||
| − | ! colspan="2" style="background:#66bbff;"|[https://parts.igem.org/Part:BBa_K404164 pCMV_VP1up_NLS_Z-EGFR-1907_(AAV2)- | + | ! colspan="2" style="background:#66bbff;"|[https://parts.igem.org/Part:BBa_K404164 pCMV_VP1up_NLS_Z-EGFR-1907_(AAV2)-VP23(ViralBrick-587KO-Empty)] |
|- | |- | ||
|'''BioBrick Nr.''' | |'''BioBrick Nr.''' | ||
| Line 38: | Line 38: | ||
VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]<br/> | VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]<br/> | ||
| − | The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal (Part:BBa_K404153, [AAV2]-NLS) (Grieger et al., 2007). | + | The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal can additionaly be inserted into VP1 (Part:BBa_K404153, [AAV2]-NLS) (Grieger et al., 2007). |
<br/> | <br/> | ||
[[Image:Freiburg10 Overview VP1Insertion.png|400px|thumb|center|Cloning scheme for insertion of motifs into VP1]]<br> | [[Image:Freiburg10 Overview VP1Insertion.png|400px|thumb|center|Cloning scheme for insertion of motifs into VP1]]<br> | ||
| Line 48: | Line 48: | ||
(BBa_K4004210)<br> | (BBa_K4004210)<br> | ||
The primary receptor of AAV-2 is the heparan sulfate proteoglycan (HSPG) receptor (Perabo et al. 2006). Its binding motif consists of five amino-acids located on the capsid surface: R484/R487, K532, R585/587. (Trepel et al. 2009). The positively charged arginine residues interact with the HSPGs' negatively charged acid residues. Opie et al. have shown that two point mutations (R585A and R588A) are sufficient to eliminate the heparin binding affinity in AAV2. (Opie et al. 2003). This ViralBrick has been created to introduce this knockout into other constructs. The biobricks with containing this knockout are annotated with „HSPG-ko“.<br> | The primary receptor of AAV-2 is the heparan sulfate proteoglycan (HSPG) receptor (Perabo et al. 2006). Its binding motif consists of five amino-acids located on the capsid surface: R484/R487, K532, R585/587. (Trepel et al. 2009). The positively charged arginine residues interact with the HSPGs' negatively charged acid residues. Opie et al. have shown that two point mutations (R585A and R588A) are sufficient to eliminate the heparin binding affinity in AAV2. (Opie et al. 2003). This ViralBrick has been created to introduce this knockout into other constructs. The biobricks with containing this knockout are annotated with „HSPG-ko“.<br> | ||
| + | <br/> | ||
| + | <h2>Characterization</h2> | ||
| + | <html> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" | ||
| + | content="text/html; charset=windows-1252"> | ||
| + | <meta name="Generator" content="Microsoft Word 12 (filtered)"> | ||
| + | <style> | ||
| + | <!-- | ||
| + | /* Font Definitions */ | ||
| + | @font-face | ||
| + | {font-family:"Cambria Math"; | ||
| + | panose-1:2 4 5 3 5 4 6 3 2 4;} | ||
| + | @font-face | ||
| + | {font-family:Cambria; | ||
| + | panose-1:2 4 5 3 5 4 6 3 2 4;} | ||
| + | @font-face | ||
| + | {font-family:Calibri; | ||
| + | panose-1:2 15 5 2 2 2 4 3 2 4;} | ||
| + | @font-face | ||
| + | {font-family:Tahoma; | ||
| + | panose-1:2 11 6 4 3 5 4 4 2 4;} | ||
| + | /* Style Definitions */ | ||
| + | p.MsoNormal, li.MsoNormal, div.MsoNormal | ||
| + | {margin-top:0cm; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:10.0pt; | ||
| + | margin-left:0cm; | ||
| + | line-height:115%; | ||
| + | font-size:11.0pt; | ||
| + | font-family:"Calibri","sans-serif";} | ||
| + | h1 | ||
| + | {mso-style-link:"Heading 1 Char"; | ||
| + | margin-top:30.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:21.6pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-21.6pt; | ||
| + | line-height:150%; | ||
| + | border:none; | ||
| + | padding:0cm; | ||
| + | font-size:16.0pt; | ||
| + | font-family:"Calibri","sans-serif";} | ||
| + | h2 | ||
| + | {mso-style-link:"Heading 2 Char"; | ||
| + | margin-top:16.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:28.8pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-28.8pt; | ||
| + | line-height:150%; | ||
| + | font-size:14.0pt; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:#002060;} | ||
| + | h3 | ||
| + | {mso-style-link:"Heading 3 Char"; | ||
| + | margin-top:16.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:36.0pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-36.0pt; | ||
| + | line-height:150%; | ||
| + | font-size:13.0pt; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:red;} | ||
| + | h4 | ||
| + | {mso-style-link:"Heading 4 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:43.2pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-43.2pt; | ||
| + | line-height:150%; | ||
| + | font-size:12.0pt; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:#00B050;} | ||
| + | h5 | ||
| + | {mso-style-link:"Heading 5 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:50.4pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-50.4pt; | ||
| + | line-height:150%; | ||
| + | font-size:11.0pt; | ||
| + | font-family:"Calibri","sans-serif";} | ||
| + | h6 | ||
| + | {mso-style-link:"Heading 6 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:4.0pt; | ||
| + | margin-left:57.6pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-57.6pt; | ||
| + | line-height:150%; | ||
| + | font-size:11.0pt; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-style:italic;} | ||
| + | p.MsoHeading7, li.MsoHeading7, div.MsoHeading7 | ||
| + | {mso-style-link:"Heading 7 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:64.8pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-64.8pt; | ||
| + | line-height:150%; | ||
| + | font-size:10.0pt; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-weight:bold; | ||
| + | font-style:italic;} | ||
| + | p.MsoHeading8, li.MsoHeading8, div.MsoHeading8 | ||
| + | {mso-style-link:"Heading 8 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:72.0pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-72.0pt; | ||
| + | line-height:150%; | ||
| + | font-size:9.0pt; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-weight:bold; | ||
| + | font-style:italic;} | ||
| + | p.MsoHeading9, li.MsoHeading9, div.MsoHeading9 | ||
| + | {mso-style-link:"Heading 9 Char"; | ||
| + | margin-top:14.0pt; | ||
| + | margin-right:0cm; | ||
| + | margin-bottom:0cm; | ||
| + | margin-left:79.2pt; | ||
| + | margin-bottom:.0001pt; | ||
| + | text-align:justify; | ||
| + | text-indent:-79.2pt; | ||
| + | line-height:150%; | ||
| + | font-size:9.0pt; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-style:italic;} | ||
| + | p.MsoAcetate, li.MsoAcetate, div.MsoAcetate | ||
| + | {mso-style-link:"Balloon Text Char"; | ||
| + | margin:0cm; | ||
| + | margin-bottom:.0001pt; | ||
| + | font-size:8.0pt; | ||
| + | font-family:"Tahoma","sans-serif";} | ||
| + | span.apple-style-span | ||
| + | {mso-style-name:apple-style-span;} | ||
| + | span.Heading1Char | ||
| + | {mso-style-name:"Heading 1 Char"; | ||
| + | mso-style-link:"Heading 1"; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | font-weight:bold;} | ||
| + | span.Heading2Char | ||
| + | {mso-style-name:"Heading 2 Char"; | ||
| + | mso-style-link:"Heading 2"; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:#002060; | ||
| + | font-weight:bold;} | ||
| + | span.Heading3Char | ||
| + | {mso-style-name:"Heading 3 Char"; | ||
| + | mso-style-link:"Heading 3"; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:red; | ||
| + | font-weight:bold;} | ||
| + | span.Heading4Char | ||
| + | {mso-style-name:"Heading 4 Char"; | ||
| + | mso-style-link:"Heading 4"; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | color:#00B050; | ||
| + | font-weight:bold;} | ||
| + | span.Heading5Char | ||
| + | {mso-style-name:"Heading 5 Char"; | ||
| + | mso-style-link:"Heading 5"; | ||
| + | font-family:"Calibri","sans-serif"; | ||
| + | font-weight:bold;} | ||
| + | span.Heading6Char | ||
| + | {mso-style-name:"Heading 6 Char"; | ||
| + | mso-style-link:"Heading 6"; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-weight:bold; | ||
| + | font-style:italic;} | ||
| + | span.Heading7Char | ||
| + | {mso-style-name:"Heading 7 Char"; | ||
| + | mso-style-link:"Heading 7"; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-weight:bold; | ||
| + | font-style:italic;} | ||
| + | span.Heading8Char | ||
| + | {mso-style-name:"Heading 8 Char"; | ||
| + | mso-style-link:"Heading 8"; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-weight:bold; | ||
| + | font-style:italic;} | ||
| + | span.Heading9Char | ||
| + | {mso-style-name:"Heading 9 Char"; | ||
| + | mso-style-link:"Heading 9"; | ||
| + | font-family:"Cambria","serif"; | ||
| + | font-style:italic;} | ||
| + | span.BalloonTextChar | ||
| + | {mso-style-name:"Balloon Text Char"; | ||
| + | mso-style-link:"Balloon Text"; | ||
| + | font-family:"Tahoma","sans-serif";} | ||
| + | .MsoPapDefault | ||
| + | {margin-bottom:10.0pt; | ||
| + | line-height:115%;} | ||
| + | @page WordSection1 | ||
| + | {size:595.3pt 841.9pt; | ||
| + | margin:70.85pt 70.85pt 2.0cm 70.85pt;} | ||
| + | div.WordSection1 | ||
| + | {page:WordSection1;} | ||
| + | /* List Definitions */ | ||
| + | ol | ||
| + | {margin-bottom:0cm;} | ||
| + | ul | ||
| + | {margin-bottom:0cm;} | ||
| + | --> | ||
| + | </style> | ||
| + | </head> | ||
| + | <body lang="DE"> | ||
| + | <div class="WordSection1"> | ||
| + | <h4><span lang="EN-US">Transduction Efficacy by Flow Cytometry</span></h4> | ||
| + | <p class="MsoNormal"><span style="font-size: 12pt; line-height: 115%;" | ||
| + | lang="EN-US">For | ||
| + | determination of transduction efficacy flow cytometry analysis was | ||
| + | conducted. 250.000 | ||
| + | AAV-293 cells were transfected with 1 µg total DNA and different ratios | ||
| + | of VP1 | ||
| + | insertion constructs in respect to the Rep/Cap(VP1KO) plasmid were | ||
| + | co-transfected. 72 hours post transfection viruses were harvested and | ||
| + | HT1080 | ||
| + | and A431 cells were transduced. 48 hours later the number of | ||
| + | transduced, or to | ||
| + | be precise mVenus positive, cells was determined via flow cytometry | ||
| + | (Fig. 1).</span></p> | ||
| + | <p style="text-align: center;" class="MsoNormal"><img | ||
| + | style="width: 597px; height: 447px;" | ||
| + | src="https://static.igem.org/mediawiki/parts/thumb/a/a1/Freiburg10_FacsAffiHT1080.png/800px-Freiburg10_FacsAffiHT1080.png" | ||
| + | alt="Image:Freiburg10 FacsAffiHT1080.png" border="0"></p> | ||
| + | <p class="MsoNormal" style="text-align: center;" align="center"><span | ||
| + | style="font-size: 12pt; line-height: 115%;"><br> | ||
| + | </span></p> | ||
| + | <p class="MsoNormal" style="text-align: center; line-height: normal;" | ||
| + | align="center"><span style="font-size: 12pt;" lang="EN-US"> </span></p> | ||
| + | <p class="MsoNormal" style="line-height: normal;"><b><span lang="EN-US">Figure | ||
| + | 1 a: | ||
| + | Flow cytometry analysis</span></b><span lang="EN-US">. Transduced and | ||
| + | therefore | ||
| + | mVenus positive HT1080 cells, infected with virus particles containing | ||
| + | Affibody | ||
| + | VP1 insertion constructs in respect to Rep/Cap(VP1KO) plasmid.</span></p> | ||
| + | <p class="MsoNormal" style="line-height: normal; text-align: center;"><span | ||
| + | lang="EN-US"></span><img style="width: 560px; height: 419px;" | ||
| + | src="https://static.igem.org/mediawiki/parts/thumb/4/48/Freiburg10_FacsAffi.png/800px-Freiburg10_FacsAffi.png" | ||
| + | alt="Image:Freiburg10 FacsAffi.png" border="0"></p> | ||
| + | <p class="MsoNormal" style="text-align: center;" align="center"><span | ||
| + | style="font-size: 12pt; line-height: 115%;"><br> | ||
| + | </span></p> | ||
| + | <p class="MsoNormal" style="text-align: center; line-height: normal;" | ||
| + | align="center"><span style="font-size: 12pt;" lang="EN-US"> </span></p> | ||
| + | <p class="MsoNormal" style="line-height: normal;"><b><span lang="EN-US">Figure | ||
| + | 1 b: | ||
| + | Flow cytometry analysis</span></b><span lang="EN-US">. Transduced and | ||
| + | therefore | ||
| + | mVenus positive A431 cells, infected with virus particles containing | ||
| + | Affibody | ||
| + | VP1 insertion constructs in respect to Rep/Cap(VP1KO) plasmid. </span></p> | ||
| + | <p class="MsoNormal" style="line-height: normal;"><span lang="EN-US"> </span></p> | ||
| + | <p class="MsoNormal"><span style="font-size: 12pt; line-height: 115%;" | ||
| + | lang="EN-US">Results | ||
| + | revealed that all virus particles remained infectious. Transduction | ||
| + | efficacy of | ||
| + | HT1080 cells was about two times less efficient in comparison to | ||
| + | unmodified | ||
| + | capsid, lipo transfected cells. While infection efficacy of A431 cells | ||
| + | significantly decreased with control viruses, the Affibody integrated | ||
| + | viruses | ||
| + | retained their targeting properties, indicating that EGFR over | ||
| + | expressing cells | ||
| + | can be specifically and efficiently infected. This also indicated that | ||
| + | VP1 | ||
| + | tolerated larger peptides inserted downstream of its unique N-terminal | ||
| + | region | ||
| + | and that this modification still allowed virus assembly and packaging.</span></p> | ||
| + | <p class="MsoNormal"><span style="font-size: 12pt; line-height: 115%;" | ||
| + | lang="EN-US"> </span></p> | ||
| + | <p class="MsoNormal"><span style="font-size: 12pt; line-height: 115%;" | ||
| + | lang="EN-US"> </span></p> | ||
| + | <p class="MsoNormal"><span style="font-size: 12pt; line-height: 115%;" | ||
| + | lang="EN-US"> </span></p> | ||
| + | <h4 style="margin-left: 0cm; text-indent: 0cm;"><span lang="EN-US"> </span></h4> | ||
| + | <h4 style="margin-left: 0cm; text-indent: 0cm;"><span lang="EN-US"> </span></h4> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 13:09, 13 January 2011
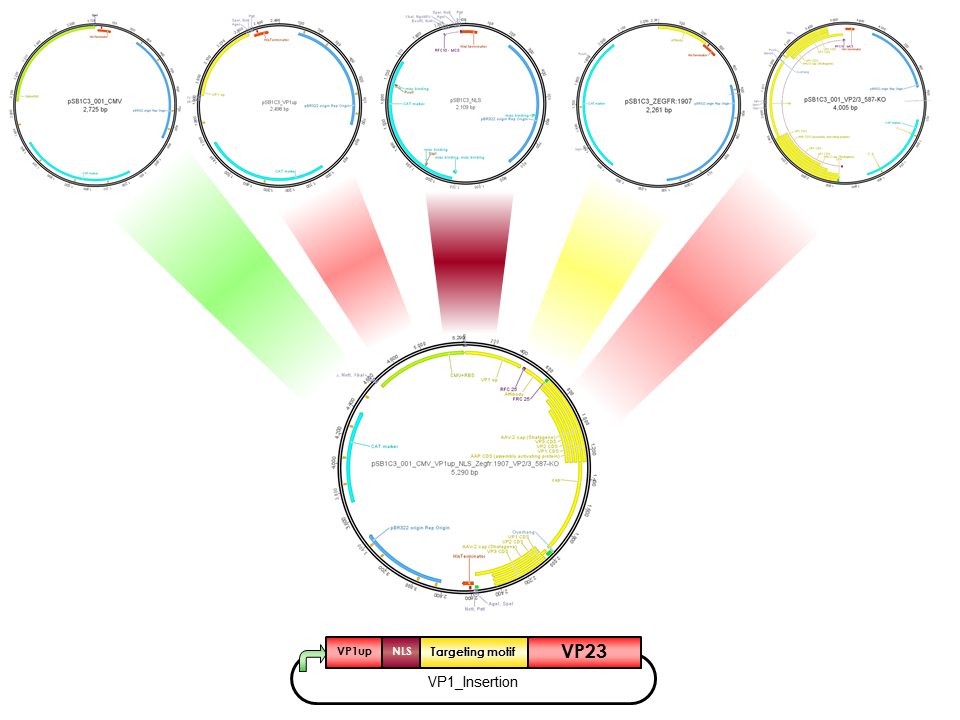
pCMV_VP1up_NLS_Z-EGFR-1907_[AAV2]-VP23 (ViralBrick-587KO-Empty)
| pCMV_VP1up_NLS_Z-EGFR-1907_(AAV2)-VP23(ViralBrick-587KO-Empty) | |
|---|---|
| BioBrick Nr. | BBa_K404164 |
| RFC standard | RFC 10 |
| Requirement | pSB1C3 |
| Source | |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
CMV
CMV promoter is derived from human Cytomegalovirus, which belongs to Herpesvirus group. All family members share the ability to remain in latent stage in the human body. CMV is located upstream of immediate-early gene. However, CMV promoter is an example of widely used promoters and is present in mammalian expression vectors. The advantage of CMV is the high-level constitutive expression in mostly all human tissues [Fitzsimons et al., 2002].
Affibody Z-EGFR-1907
(BBa_K404302)
Affibodies are small (6 kDa), soluble high-affinity proteins. They are derived from the IgG-binding B domain of the Staphylococcal protein A, which was engineered to specifically bind to certain peptides or proteins. This so-called Z domain consists of an antiparallel three-helix bundle and is advantageous due to its proteolytic and thermodynamic stability, its good folding properties and the ease of production via recombinant bacteria (Nord et al. 1997). Affibodies can be used for example for tumor targeting (Wikman et al. 2004) and diagnostic imaging applications(Friedman et al. 2008)(Orlova et al. 2007). The ZEGFR:1907 Affibody was engineered to specifically bind the EGF receptor with an affinity determined to be KD = 2.8 nM (Friedman et al. 2008).
The EGF receptor is overexpressed in certain types of tumors, e.g. in breast (Walker and Dearing 1999), lung (Hirsch et al. 2003) and bladder (Colquhoun and Mellon 2002) carcinomas, and is therefore a suitable target for cancer imaging or therapeutic applications. Because of their good tumor uptake, and their property to become internalized into the target cells with an efficiency of 19 – 24% within one hour – compared to 45% of the natural ligand EGF - the ZEGFR:1907 Affibody was chosen for therapeutic applications by the Freiburg iGEM Team 2010 (Göstring et al. 2010; Friedman et al. 2008).
Capsid
(BBa_K404006)
The AAV capsid consists of 60 capsid protein subunits composed of the three cap proteins VP1, VP2, and VP3, which are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of the VP proteins. VP1, VP2 and VP3 share a common C terminus and stop codon, but begin with a different start codon. The N termini of VP1 and VP2 play important roles in infection and contain motifs that are highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (NLSs). These elements are conserved in almost all parvoviruses. (Johnson et al., 2010a).
Whereas VP1 is translated from the minor spliced mRNA, while VP2 and VP3 are translated from the major spliced mRNA. The minor spliced product is approximately 10-fold less abundant than the major spliced mRNA. Thus, there is much less VP1 than VP2 and VP3 resulting in a capsid stoichiometric ratio of 1:1:10. The N terminus of VP1 has an extension of 65 amino acids including an additional extension of 138 N-terminal amino acids forming the unique portion of VP1. It contains a motif of about 70 amino acids that is highly homologous to phospholipase A2 (PLA2) domain. Furthermore, there are nuclear localization sequences (BR)(+), which are supposed to be necessary for endosomal escape and nuclear entry. (Bleker, Pawlita, & Kleinschmidt, 2006), (DiPrimio, Asokan, Govindasamy, Agbandje-McKenna, & Samulski, 2008), (Johnson et al., 2010a)
VP1up
VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]
The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal can additionaly be inserted into VP1 (Part:BBa_K404153, [AAV2]-NLS) (Grieger et al., 2007).
NLS
NLS are located in basic regions on the N terminus of VP2 (35 aa) and VP1 (172 aa) and mediate genome delivery into the nucleus and transduction [Hoque et al.,1999; Grieger et al., 2006]. Nuclear localisation sequence is hydrophilic and contains ß-turn and coil regions [Kalderon, et al, 1984]. It was also described in CPV and MVM viruses. Compared to CPV, MVM virus contains several NLS within the capsid, which are activated at different infection stages [Lombardo et al., 2000; Lombardo et al., 2002]
ViralBrick 587-KO empty
(BBa_K4004210)
The primary receptor of AAV-2 is the heparan sulfate proteoglycan (HSPG) receptor (Perabo et al. 2006). Its binding motif consists of five amino-acids located on the capsid surface: R484/R487, K532, R585/587. (Trepel et al. 2009). The positively charged arginine residues interact with the HSPGs' negatively charged acid residues. Opie et al. have shown that two point mutations (R585A and R588A) are sufficient to eliminate the heparin binding affinity in AAV2. (Opie et al. 2003). This ViralBrick has been created to introduce this knockout into other constructs. The biobricks with containing this knockout are annotated with „HSPG-ko“.
Characterization
Transduction Efficacy by Flow Cytometry
For determination of transduction efficacy flow cytometry analysis was conducted. 250.000 AAV-293 cells were transfected with 1 µg total DNA and different ratios of VP1 insertion constructs in respect to the Rep/Cap(VP1KO) plasmid were co-transfected. 72 hours post transfection viruses were harvested and HT1080 and A431 cells were transduced. 48 hours later the number of transduced, or to be precise mVenus positive, cells was determined via flow cytometry (Fig. 1).

Figure 1 a: Flow cytometry analysis. Transduced and therefore mVenus positive HT1080 cells, infected with virus particles containing Affibody VP1 insertion constructs in respect to Rep/Cap(VP1KO) plasmid.

Figure 1 b: Flow cytometry analysis. Transduced and therefore mVenus positive A431 cells, infected with virus particles containing Affibody VP1 insertion constructs in respect to Rep/Cap(VP1KO) plasmid.
Results revealed that all virus particles remained infectious. Transduction efficacy of HT1080 cells was about two times less efficient in comparison to unmodified capsid, lipo transfected cells. While infection efficacy of A431 cells significantly decreased with control viruses, the Affibody integrated viruses retained their targeting properties, indicating that EGFR over expressing cells can be specifically and efficiently infected. This also indicated that VP1 tolerated larger peptides inserted downstream of its unique N-terminal region and that this modification still allowed virus assembly and packaging.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2609
Illegal XhoI site found at 698
Illegal XhoI site found at 884 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 665
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 3135
Illegal SapI site found at 2046
References
Mellon. 2002. Epidermal growth factor receptor and bladder cancer.Postgraduate
medical journal78, no. 924 (October): 584-9.
doi:10.1136/pmj.78.924.584.
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1742539&tool=pmcentrez&rendertype=abstract.
Friedman,
Mikaela, Anna
Orlova, Eva Johansson, Tove L J Eriksson, Ingmarie Höidén-Guthenberg,
Vladimir
Tolmachev, Fredrik Y Nilsson, and Stefan Ståhl. 2008. Directed
evolution to low
nanomolar affinity of a tumor-targeting epidermal growth factor
receptor-binding affibody molecule. Journal of molecular
biology376,
no. 5: 1388-402. doi:10.1016/j.jmb.2007.12.060.
http://www.ncbi.nlm.nih.gov/pubmed/18207161.
Göstring,
Lovisa, Ming Tsuey
Chew, Anna Orlova, Ingmarie Höidén-guthenberg, Anders Wennborg, Jörgen
Carlsson, and Fredrik Y Frejd. 2010. Quantification of internalization
of
EGFR-binding Affibody molecules: Methodological aspects. International
Journal of Oncology 36, no. 4 (March): 757-763.
doi:10.3892/ijo_00000551.
http://www.spandidos-publications.com/ijo/36/4/757.
Hirsch,Fred R, Marileila Varella-Garcia, Paul a Bunn, Michael V Di Maria, Robert Veve, Roy M Bremmes,
Anna E Barón, Chan Zeng, and Wilbur a Franklin. 2003. Epidermal growth factor
receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology
21, no. 20 (October): 3798-807. doi:10.1200/JCO.2003.11.069.
http://www.ncbi.nlm.nih.gov/pubmed/12953099.
Nord, K, E Gunneriusson, J Ringdahl, S Ståhl, M Uhlén, and P A Nygren. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nature biotechnology 15, no. 8 (August): 772-7. doi:10.1038/nbt0897-772. http://www.ncbi.nlm.nih.gov/pubmed/9255793.
Orlova,
Anna, Vladimir
Tolmachev, Rikard Pehrson, Malin Lindborg, Thuy Tran, Mattias
Sandström,
Fredrik Y Nilsson, Anders Wennborg, Lars Abrahmsén, and Joachim
Feldwisch.
2007. Synthetic affibody molecules: a novel class of affinity ligands
for
molecular imaging of HER2-expressing malignant tumors. Cancer
research
67, no. 5 (March): 2178-86. doi:10.1158/0008-5472.CAN-06-2887.
http://www.ncbi.nlm.nih.gov/pubmed/17332348.
Walker,
R a, and S J Dearing.
1999. Expression of epidermal growth factor receptor mRNA and protein
in
primary breast carcinomas. Breast cancer research and
treatment53, no.
2 (January): 167-76. http://www.ncbi.nlm.nih.gov/pubmed/10326794.
Wikman,
M, a-C Steffen, E
Gunneriusson, V Tolmachev, G P Adams, J Carlsson, and S Ståhl. 2004.
Selection
and characterization of HER2/neu-binding affibody ligands. Protein
engineering, design & selection : PEDS 17, no. 5
(May): 455-62.
doi:10.1093/protein/gzh053. http://www.ncbi.nlm.nih.gov/pubmed/15208403.
Bleker, S., Pawlita, M., & Kleinschmidt, J., 2006. IImpact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. Journal of virology, 80(2), 810-820. doi: 10.1128/JVI.80.2.810.
DiPrimio, N., Asokan, A., Govindasamy, L., Agbandje-McKenna, M., & Samulski, R. J. , 2008. Surface loop dynamics in adeno-associated virus capsid assembly. Journal of virology, 82(11), 5178-89. doi: 10.1128/JVI.02721-07.
Hoque, 1999. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. Journal of Virology, 73(9), pp.7912-7915.
Kalderon, D, 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature, 311(5981), pp.33-38
Johnson, J.S., 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. Journal of virology, 84(17), 8888-902. doi: 10.1128/JVI.00687-10.
Lombardo, E, 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. Journal of Virology, 74(8), pp.3804-3814
Lombardo, E , 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. Journal of Virology, 76(14), pp.7049-7059
Bleker, S., Pawlita, M., & Kleinschmidt, J., 2006. IImpact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. Journal of virology, 80(2), 810-820. doi: 10.1128/JVI.80.2.810.
Canaan, 2004. Interfacial enzymology of parvovirus phospholipases A2. The Journal of Biological Chemistry, 279(15), pp.14502-14508.
DiPrimio, N., Asokan, A., Govindasamy, L., Agbandje-McKenna, M., & Samulski, R. J. , 2008. Surface loop dynamics in adeno-associated virus capsid assembly. Journal of virology, 82(11), 5178-89. doi: 10.1128/JVI.02721-07.
Girod, A., 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. The Journal of general virology, 83(Pt 5), pp.973-978. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11961250.
Johnson, J.S., 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. Journal of virology, 84(17), 8888-902. doi: 10.1128/JVI.00687-10.
Zadori, Z , 2001. A viral phospholipase A2 is required for parvovirus infectivity. Developmental Cell, 1(2), pp.291-302. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11702787.
Grieger et al., (2007). Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J Virol. 2007 Aug;81(15):7833-43. Epub 2007 May 16.