Difference between revisions of "Part:BBa K346001"
Spring zhq (Talk | contribs) |
|||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
[[Image:MerR-structure.jpg|center]] | [[Image:MerR-structure.jpg|center]] | ||
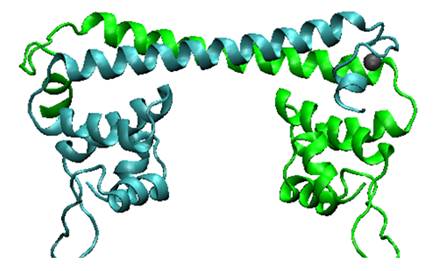
| − | '''Fig.1. Predicted MerR dimer structure.''' | + | '''Fig.1. Predicted MerR dimer structure.''' The predicted three dimensional conformation of Hg (II) bound MerR structure by our 3D modeling. Two monomers are indicated in green and cyan, respectively. MerR can be divided into two modular domains: the metal binding domain at the C-terminal and DNA binding domain at the N-terminal. The signature of MerR family is a helix-turn-helix (HTH) motif followed by a coiled-coil region, namely, a similar N-terminal helix-turn-helix DNA binding domains and a C-terminal effector binding domains that are specific to the effector (eg. metal ions)to be recognized |
---- | ---- | ||
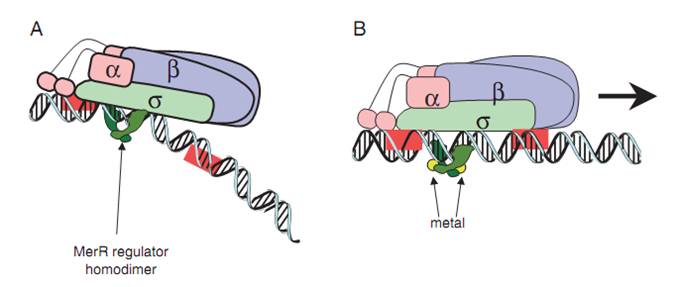
| − | + | MerR protein, in the form of homodimer, binds to a dyad sequence between the -10 element and -35 region of element PmerT. The binding, however, is independent of the presence of Hg (II). When the apo-MerR dimer binds to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has an unusually long spacer of 19 bp, the contact of RNA polymerase is sloppy. When Hg (II) is available in the environment, ions bind to the interface of MerR dimer. The Hg-bound MerR dimer can result in a structural distortion of PmerT, allowing the RNA polymerase tight contacts to be made, leading to the expression of down-stream genes (Fig.2). | |
[[Image:MerR-dimer.jpg|center]] | [[Image:MerR-dimer.jpg|center]] | ||
| − | '''Fig.2. | + | '''Fig.2. The mechanism of MerR transcription activation.''' A: The dimeric MerR regulator binds to the operator region of PmerT promoter and recruits RNA polymerase, forming a ternary complex. Transcription is slightly repressed because the apo-MerR regulator dimer has bent the promoter DNA such that RNA polymerase does not contact it properly. B: Upon binding the cognate metal ions, the metallated MerR homodimer causes a realignment of the promoter such that RNA polymerase contacts the -35 and -10 sequences leading to open complex formation and transcription. Adapted from Brown et al. |
| − | < | + | ===BEAS_China 2019: Experimental Characterization=== |
| − | === | + | <html> |
| + | <p><b>BEAS_China 2019 used BBa_K3143675 in our basic mercury sensor design: J23 family-merR-pMerR-sfGFP-terminator. </b>This sensor has a constitutive promoter (J23 family) that drives the expression of an mercury receptor MerR, which would de-repress its cognate promoter merR on murcury binding and trigger the expression of a reporter gene, gfp. </p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/b/b1/T--BEAS_China--Description_Basic_sensor_Principle.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Figure 3: The scheme of basic sensor design. </h6> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <p>We select three constitutive promoters of varying strengths from iGEM promoter library (Fig. 4A). The sensors were then compared under various HgNO3 induction conditions (Fig. 4B). The results showed that the weaker the promoter (that is, the lower the MerR receptor concentration), the more sensitive and higher the dynamic range of the sensor.</p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/6/6c/T--BEAS_China--Demonstration_Fig_1a_%26_1b.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Figure 4: <strong>A</strong> Different constitutively J23 family promoter measured strength (Data source: iGEM) <strong>B</strong> Tuning mercury receptor merR’s intracellular density by varying the strength of J23 prmoter </h6> | ||
| + | </div> | ||
| + | |||
| + | <p>We fitted the sensors’ dose–response curves to a Hill function-based biochemical model to describe their input-output relationships. (Fig 5 and Table 1) </p> | ||
| + | <ul> | ||
| + | <li> | ||
| + | <p>The Hill constant EC50 is the inducer concentration that provokes half-maximal activation of a sensor; EC50 is negatively correlated with sensitivity.</p> | ||
| + | </li> | ||
| + | <li> | ||
| + | <p>KTop is the sensor’s maximum output expression level; KTop is positively correlated with output amplitude.</p> | ||
| + | </li> | ||
| + | </ul> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/d/d0/T--BEAS_China--Demonstration_Fig_2.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Figure 5: The equation used to fit the sensors’ dose–response curves to a Hill function based biochemical model to describe their input-GFPput relationships | ||
| + | </h6> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/2a/T--BEAS_China--Demonstration_Table1.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Table 1: Best fits for the characterized response of the various sensors circuits in this study | ||
| + | </h6> | ||
| + | </div> | ||
| + | <p>Here, EC50 showed a 2.7-fold decrease and KTop showed a 3.5-fold increase from high to low MerR levels (Fig. 6a & 6b ), confirming that the mercury sensor’s sensitivity and output amplitude were both increased while the MerR intracellular concentration was decreased. </p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/5/56/T--BEAS_China--Demonstration_Fig_3A_%26_3B.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Figure 6: The maximum output (KTop) and EC50 of the sensor’s dose response against the relevant intracellular MerR concentrations </h6> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
<!-- --> | <!-- --> | ||
Latest revision as of 14:38, 21 October 2019
RBS (B0034) + MerR (mercury-responsive transcription factor)
This part was designed as a translational unit for MerR expression.
The mercury resistance operon, mer, enables bacteria to avoid and remove toxic metal Hg, under the regulation of the MerR family transcriptional factor MerR. MerR acts as an activator of mer genes in response to the presence of Hg (II), while it will turn into a weak repressor in the absence of Hg (II), to maintain its own expression at certain level.
Fig.1. Predicted MerR dimer structure. The predicted three dimensional conformation of Hg (II) bound MerR structure by our 3D modeling. Two monomers are indicated in green and cyan, respectively. MerR can be divided into two modular domains: the metal binding domain at the C-terminal and DNA binding domain at the N-terminal. The signature of MerR family is a helix-turn-helix (HTH) motif followed by a coiled-coil region, namely, a similar N-terminal helix-turn-helix DNA binding domains and a C-terminal effector binding domains that are specific to the effector (eg. metal ions)to be recognized
MerR protein, in the form of homodimer, binds to a dyad sequence between the -10 element and -35 region of element PmerT. The binding, however, is independent of the presence of Hg (II). When the apo-MerR dimer binds to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has an unusually long spacer of 19 bp, the contact of RNA polymerase is sloppy. When Hg (II) is available in the environment, ions bind to the interface of MerR dimer. The Hg-bound MerR dimer can result in a structural distortion of PmerT, allowing the RNA polymerase tight contacts to be made, leading to the expression of down-stream genes (Fig.2).
Fig.2. The mechanism of MerR transcription activation. A: The dimeric MerR regulator binds to the operator region of PmerT promoter and recruits RNA polymerase, forming a ternary complex. Transcription is slightly repressed because the apo-MerR regulator dimer has bent the promoter DNA such that RNA polymerase does not contact it properly. B: Upon binding the cognate metal ions, the metallated MerR homodimer causes a realignment of the promoter such that RNA polymerase contacts the -35 and -10 sequences leading to open complex formation and transcription. Adapted from Brown et al.
BEAS_China 2019: Experimental Characterization
BEAS_China 2019 used BBa_K3143675 in our basic mercury sensor design: J23 family-merR-pMerR-sfGFP-terminator. This sensor has a constitutive promoter (J23 family) that drives the expression of an mercury receptor MerR, which would de-repress its cognate promoter merR on murcury binding and trigger the expression of a reporter gene, gfp.

Figure 3: The scheme of basic sensor design.
We select three constitutive promoters of varying strengths from iGEM promoter library (Fig. 4A). The sensors were then compared under various HgNO3 induction conditions (Fig. 4B). The results showed that the weaker the promoter (that is, the lower the MerR receptor concentration), the more sensitive and higher the dynamic range of the sensor.

Figure 4: A Different constitutively J23 family promoter measured strength (Data source: iGEM) B Tuning mercury receptor merR’s intracellular density by varying the strength of J23 prmoter
We fitted the sensors’ dose–response curves to a Hill function-based biochemical model to describe their input-output relationships. (Fig 5 and Table 1)
-
The Hill constant EC50 is the inducer concentration that provokes half-maximal activation of a sensor; EC50 is negatively correlated with sensitivity.
-
KTop is the sensor’s maximum output expression level; KTop is positively correlated with output amplitude.

Figure 5: The equation used to fit the sensors’ dose–response curves to a Hill function based biochemical model to describe their input-GFPput relationships

Table 1: Best fits for the characterized response of the various sensors circuits in this study
Here, EC50 showed a 2.7-fold decrease and KTop showed a 3.5-fold increase from high to low MerR levels (Fig. 6a & 6b ), confirming that the mercury sensor’s sensitivity and output amplitude were both increased while the MerR intracellular concentration was decreased.

Figure 6: The maximum output (KTop) and EC50 of the sensor’s dose response against the relevant intracellular MerR concentrations
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]