Difference between revisions of "Part:BBa K404152"
| (3 intermediate revisions by one other user not shown) | |||
| Line 20: | Line 20: | ||
|[http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] | |[http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] | ||
|} | |} | ||
| + | [[Image:Freiburg10 VP1upSchema.png|thumb|200px|right]] | ||
<br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
<h3>General informations</h3> | <h3>General informations</h3> | ||
| − | The AAV capsid consists of 60 capsid protein subunits composed of the three cap proteins VP1, VP2, and VP3, which are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of the VP proteins. VP1, VP2 and VP3 share a common C terminus and stop codon, but begin with a different start codon. The N | + | The AAV capsid consists of 60 capsid protein subunits composed of the three cap proteins VP1, VP2, and VP3, which are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of the VP proteins. VP1, VP2 and VP3 share a common C terminus and stop codon, but begin with a different start codon. The N-terminus of VP1 plays an important role in infection and contains a motif highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+). VP 2 contains basic regions, too. These elements are conserved in almost all parvoviruses. (Johnson et al., 2010a). <br/> |
<br/> | <br/> | ||
Whereas VP1 is translated from the minor spliced mRNA, while VP2 and VP3 are translated from the major spliced mRNA. The minor spliced product is approximately 10-fold less abundant than the major spliced mRNA. Thus, there is much less VP1 than VP2 and VP3 resulting in a capsid stoichiometric ratio of 1:1:10. The N terminus of VP1 has an extension of 65 amino acids including an additional extension of 138 N-terminal amino acids forming the unique portion of VP1. It contains a motif of about 70 amino acids that is highly homologous to phospholipase A2 (PLA2) domain. Furthermore, there are nuclear localization sequences (BR)(+), which are supposed to be necessary for endosomal escape and nuclear entry. (Bleker, Pawlita, & Kleinschmidt, 2006), (DiPrimio, Asokan, Govindasamy, Agbandje-McKenna, & Samulski, 2008), (Johnson et al., 2010a) | Whereas VP1 is translated from the minor spliced mRNA, while VP2 and VP3 are translated from the major spliced mRNA. The minor spliced product is approximately 10-fold less abundant than the major spliced mRNA. Thus, there is much less VP1 than VP2 and VP3 resulting in a capsid stoichiometric ratio of 1:1:10. The N terminus of VP1 has an extension of 65 amino acids including an additional extension of 138 N-terminal amino acids forming the unique portion of VP1. It contains a motif of about 70 amino acids that is highly homologous to phospholipase A2 (PLA2) domain. Furthermore, there are nuclear localization sequences (BR)(+), which are supposed to be necessary for endosomal escape and nuclear entry. (Bleker, Pawlita, & Kleinschmidt, 2006), (DiPrimio, Asokan, Govindasamy, Agbandje-McKenna, & Samulski, 2008), (Johnson et al., 2010a) | ||
| Line 29: | Line 30: | ||
VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]<br/> | VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]<br/> | ||
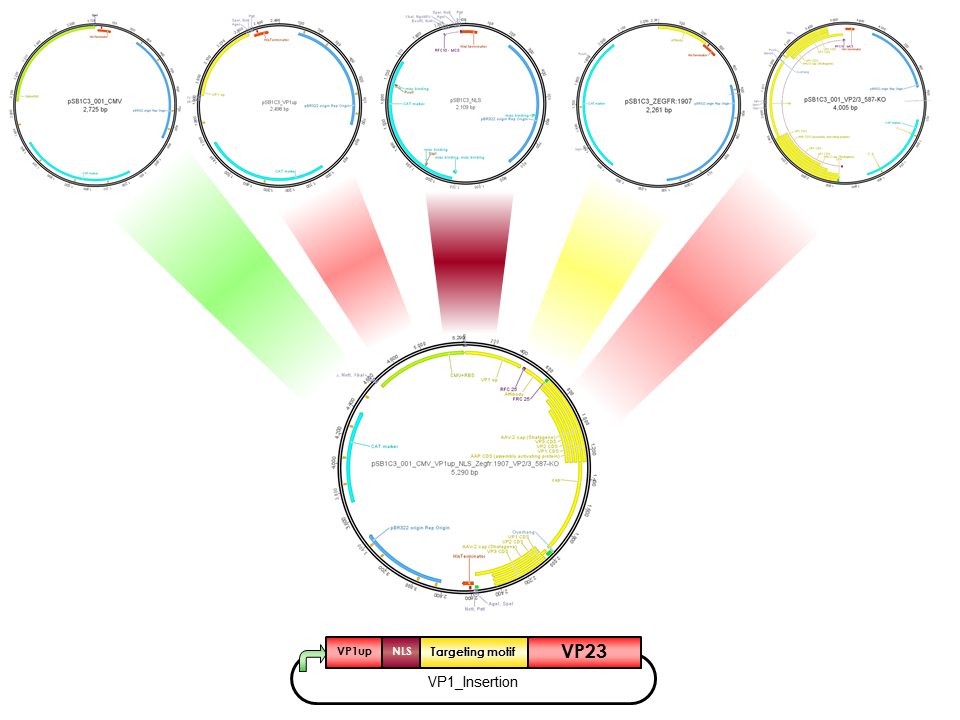
| − | The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal (Part:BBa_K404153, [AAV2]-NLS). | + | The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal can additionaly be inserted into VP1(Part:BBa_K404153, [AAV2]-NLS) (Grieger et al., 2007). |
| + | <br/> | ||
| + | [[Image:Freiburg10 Overview VP1Insertion.png|400px|thumb|center|Cloning scheme for insertion of motifs into VP1]] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 50: | Line 53: | ||
<b>Johnson, J.S.</b>, 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. Journal of virology, 84(17), 8888-902. doi: 10.1128/JVI.00687-10.<br /> | <b>Johnson, J.S.</b>, 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. Journal of virology, 84(17), 8888-902. doi: 10.1128/JVI.00687-10.<br /> | ||
<b>Zadori, Z </b>, 2001. A viral phospholipase A2 is required for parvovirus infectivity. Developmental Cell, 1(2), pp.291-302. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11702787. <br /> | <b>Zadori, Z </b>, 2001. A viral phospholipase A2 is required for parvovirus infectivity. Developmental Cell, 1(2), pp.291-302. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11702787. <br /> | ||
| + | <b>Grieger et al.</b>, (2007). Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J Virol. 2007 Aug;81(15):7833-43. Epub 2007 May 16. | ||
Latest revision as of 15:11, 31 October 2010
[AAV2]-VP1up
| [AAV2-VP1up] | |
|---|---|
| BioBrick Nr. | BBa_K404152 |
| RFC standard | RFC 25 |
| Requirement | pSB1C3 |
| Source | |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
General informations
The AAV capsid consists of 60 capsid protein subunits composed of the three cap proteins VP1, VP2, and VP3, which are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of the VP proteins. VP1, VP2 and VP3 share a common C terminus and stop codon, but begin with a different start codon. The N-terminus of VP1 plays an important role in infection and contains a motif highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+). VP 2 contains basic regions, too. These elements are conserved in almost all parvoviruses. (Johnson et al., 2010a).
Whereas VP1 is translated from the minor spliced mRNA, while VP2 and VP3 are translated from the major spliced mRNA. The minor spliced product is approximately 10-fold less abundant than the major spliced mRNA. Thus, there is much less VP1 than VP2 and VP3 resulting in a capsid stoichiometric ratio of 1:1:10. The N terminus of VP1 has an extension of 65 amino acids including an additional extension of 138 N-terminal amino acids forming the unique portion of VP1. It contains a motif of about 70 amino acids that is highly homologous to phospholipase A2 (PLA2) domain. Furthermore, there are nuclear localization sequences (BR)(+), which are supposed to be necessary for endosomal escape and nuclear entry. (Bleker, Pawlita, & Kleinschmidt, 2006), (DiPrimio, Asokan, Govindasamy, Agbandje-McKenna, & Samulski, 2008), (Johnson et al., 2010a)
VP1up
VP1up protein is derived from the unique N-terminal region of VP1 protein. It contains a Phospholipase A2 motif which is essential for successful infection [Canaan et al., 2004; Zadori et al., 2001; Girod et al., 2002]
The Freiburg iGEM Team 2010 created this part in order integrate motifs, which are desired to be surface exposed, into the VP1 open reading frame. For this purpose VP1up needs to be fused to the N-terminus of the motif of interest, followed by coupling the resulting construct to VP2/3 (BBa_K404150, [AAV2]-VP23). For better infectivity a nuclear localization signal can additionaly be inserted into VP1(Part:BBa_K404153, [AAV2]-NLS) (Grieger et al., 2007).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 28
Illegal XhoI site found at 214 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
Bleker, S., Pawlita, M., & Kleinschmidt, J., 2006. IImpact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. Journal of virology, 80(2), 810-820. doi: 10.1128/JVI.80.2.810.
Canaan, 2004. Interfacial enzymology of parvovirus phospholipases A2. The Journal of Biological Chemistry, 279(15), pp.14502-14508.
DiPrimio, N., Asokan, A., Govindasamy, L., Agbandje-McKenna, M., & Samulski, R. J. , 2008. Surface loop dynamics in adeno-associated virus capsid assembly. Journal of virology, 82(11), 5178-89. doi: 10.1128/JVI.02721-07.
Girod, A., 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. The Journal of general virology, 83(Pt 5), pp.973-978. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11961250.
Johnson, J.S., 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. Journal of virology, 84(17), 8888-902. doi: 10.1128/JVI.00687-10.
Zadori, Z , 2001. A viral phospholipase A2 is required for parvovirus infectivity. Developmental Cell, 1(2), pp.291-302. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11702787.
Grieger et al., (2007). Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J Virol. 2007 Aug;81(15):7833-43. Epub 2007 May 16.