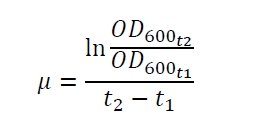Difference between revisions of "Part:BBa K389004:Experience"
(→mRFP vs. luciferase as reporter gene) |
|||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | __TOC__ | |
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
how you used this part and how it worked out. | how you used this part and how it worked out. | ||
| Line 6: | Line 6: | ||
====Accumulation of luciferase==== | ====Accumulation of luciferase==== | ||
| − | The production of the reporter gene luciferase by ''Escherichia coli'' is dependent from the specific growth rate µ (fig. 1A) and the growth phase (fig. 1B). Fig. 1A shows the specific production rate q<sub>P</sub> which was determined with equation (1) | + | The production of the reporter gene luciferase by ''Escherichia coli'' is dependent from the specific growth rate µ (fig. 1A, eq. (2)) and the growth phase (fig. 1B). Fig. 1A shows the specific production rate q<sub>P</sub> which was determined with equation (1) |
| − | [[Image:Bielefeld_Luc_qP.jpg| | + | [[Image:Bielefeld_Luc_qP.jpg|330px|center]] <div align="right">(1)</div> |
| − | plotted against the specific growth rate µ which was determined with equation (2) | + | with the amount of product P, the cell count X and the relative luminescence units RLU plotted against the specific growth rate µ which was determined with equation (2) |
| − | [[Image:. | + | [[Image:Bielefeld_Specific_growth_rate.jpg|190px|center]]. <div align="right">(2)</div> |
| Line 21: | Line 21: | ||
| − | [[Image:Bielefeld_Luc_bildung_zu_wachstum.jpg| | + | [[Image:Bielefeld_Luc_bildung_zu_wachstum.jpg|750px|thumb|center|'''Fig. 1A: Specific production rates q<sub>P</sub> of many different cultivations of <partinfo>K389015</partinfo> in ''E. coli'' DB3.1, some induced with acetosyringone, some uninduced. Fig. 1B: Cultivation of <partinfo>K389307</partinfo> in ''E. coli'' TOP10.''']] |
| + | |||
| + | ====Kinetic of luciferin conversion==== | ||
| + | Look up the general luminescence measurement protocol [[Part:BBa_K389015:Experience#Measurement protocol | here]]. | ||
| + | |||
| + | To determine the ideal time of RLU measurement after adding luciferin to the cell lysate a kinetic of this reaction was recorded with the luminometer (fig. 2). A maximum is seen between 20 - 40 s. So a measurement with a delay of at least 20 s from the time luciferin is added to the cell lysate is recommended. In addition, the time between adding the luciferin to the cell lysate and the measurement should not be more than 40 s. | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_Luc_kinetik.jpg|600px|thumb|center|'''Fig. 2: Kinetic of the luciferase reaction after adding luciferin to a cell lysate.''']] | ||
| + | |||
| + | |||
| + | ====Sensitivity==== | ||
| + | To determine the sensitivity of the reporter gene luciferase the limit of detection (LOD) and the limit of quantification (LOQ) are calculated. For this calculation the noise (standard derivation s<sub>R</sub>) and the arithmetic mean (x<sub>R</sub>) of the reference measurement, so cells without luciferase gene, are collected. Afterwards the LOD is calculated with equation (3) and the LOQ with equation (4): | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_LOD.jpg|130px|center]] <div align="right">(3)</div> | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_LOQ.jpg|130px|center]] <div align="right">(4)</div> | ||
| + | |||
| + | |||
| + | The LOD is 162 RLU and the LOQ is 306 RLU. This equals 0.3 % and 0.7 %, respectively, of the maximal RLU measured with a luciferase gene under the control of the weak constitutive promoter <partinfo>J23103</partinfo> in <partinfo>pSB1A2</partinfo> (measured with part <partinfo>K389302</partinfo> in <partinfo>pSB1A2</partinfo>). So the luciferase is a very sensitive reporter gene. | ||
| + | |||
| + | |||
| + | ====mRFP vs. luciferase as reporter gene==== | ||
| + | In the following table a short comparison between luciferase and mRFP as reporter genes for promoter activity is shown. A (+) means high and a (-) means low. Click on a (+) or (-) in the table to see the adequate results. | ||
| + | |||
| + | <center> | ||
| + | Table 1: Comparison between luciferase and mRFP as reporter genes. | ||
| + | |||
| + | {|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | ||
| + | |- | ||
| + | !style="border-style: solid; border-width: 1px"| | ||
| + | !style="border-style: solid; border-width: 1px"| Luciferase | ||
| + | !style="border-style: solid; border-width: 1px"| mRFP | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Sensitivity | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#sensitivity | (+) ]] | ||
| + | |style="border-style: solid; border-width: 1px"| (-) | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Stability | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Accumulation of luciferase | (-)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [https://parts.igem.org/PBAD_Promoter_Family#Expression_Rate (+)] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Response time | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389015:Experience#Response time | (+)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [https://parts.igem.org/PBAD_Promoter_Family#Expression_Rate (-)] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Degradation | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Accumulation of luciferase | (+)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [https://parts.igem.org/PBAD_Promoter_Family#Expression_Rate (-)] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Reproducibility | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389015:Experience#Data Analysis | (-)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389016:Experience#Data analysis for BBa_K389016 | (+)]] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Growth dependent | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Accumulation of luciferase | (+)]] | ||
| + | |style="border-style: solid; border-width: 1px"| (+) | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Costs / work | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Measurement Protocols | (+)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389016:Experience#Protocols | (-)]] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Sample volume | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Measurement Protocols | (-)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389016:Experience#Protocols | (+)]] | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| Online measuring | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_K389004:Experience#Measurement Protocols | (-)]] | ||
| + | |style="border-style: solid; border-width: 1px"| [[Part:BBa_F2620:Experience#Growth_conditions | (+)]] | ||
| + | |} | ||
| + | </center> | ||
====Measurement protocol==== | ====Measurement protocol==== | ||
| Line 33: | Line 105: | ||
* For the measurement thaw by placing the tubes in room temperature water | * For the measurement thaw by placing the tubes in room temperature water | ||
| − | * Add 300 µL of freshly prepared lysis mix ( | + | * Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL<sup>-1</sup> lysozyme, 2.5 mg mL<sup>-1</sup> BSA, add water for desired volume) |
* Mix and incubate the cells for 10 minutes at room temperature | * Mix and incubate the cells for 10 minutes at room temperature | ||
| Line 45: | Line 117: | ||
** Delay: 20 secs | ** Delay: 20 secs | ||
** Integration: 3 secs | ** Integration: 3 secs | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | http://www.promega.com/tbs/tb281/tb281.pdf | ||
Latest revision as of 14:57, 26 October 2010
Contents
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K389004
Accumulation of luciferase
The production of the reporter gene luciferase by Escherichia coli is dependent from the specific growth rate µ (fig. 1A, eq. (2)) and the growth phase (fig. 1B). Fig. 1A shows the specific production rate qP which was determined with equation (1)
with the amount of product P, the cell count X and the relative luminescence units RLU plotted against the specific growth rate µ which was determined with equation (2)
With decreasing growth rate the luciferase production slows down and with stopping growth the luciferase is degraded. Especially in the stationary growth phase the luciferase degradation is high (fig. 1B).

Kinetic of luciferin conversion
Look up the general luminescence measurement protocol here.
To determine the ideal time of RLU measurement after adding luciferin to the cell lysate a kinetic of this reaction was recorded with the luminometer (fig. 2). A maximum is seen between 20 - 40 s. So a measurement with a delay of at least 20 s from the time luciferin is added to the cell lysate is recommended. In addition, the time between adding the luciferin to the cell lysate and the measurement should not be more than 40 s.
Sensitivity
To determine the sensitivity of the reporter gene luciferase the limit of detection (LOD) and the limit of quantification (LOQ) are calculated. For this calculation the noise (standard derivation sR) and the arithmetic mean (xR) of the reference measurement, so cells without luciferase gene, are collected. Afterwards the LOD is calculated with equation (3) and the LOQ with equation (4):
The LOD is 162 RLU and the LOQ is 306 RLU. This equals 0.3 % and 0.7 %, respectively, of the maximal RLU measured with a luciferase gene under the control of the weak constitutive promoter BBa_J23103 in pSB1A2 (measured with part BBa_K389302 in pSB1A2). So the luciferase is a very sensitive reporter gene.
mRFP vs. luciferase as reporter gene
In the following table a short comparison between luciferase and mRFP as reporter genes for promoter activity is shown. A (+) means high and a (-) means low. Click on a (+) or (-) in the table to see the adequate results.
Table 1: Comparison between luciferase and mRFP as reporter genes.
| Luciferase | mRFP | |
|---|---|---|
| Sensitivity | (+) | (-) |
| Stability | (-) | (+) |
| Response time | (+) | (-) |
| Degradation | (+) | (-) |
| Reproducibility | (-) | (+) |
| Growth dependent | (+) | (+) |
| Costs / work | (+) | (-) |
| Sample volume | (-) | (+) |
| Online measuring | (-) | (+) |
Measurement protocol
- For the luciferase detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer
- Prepare reaction tubes with 10 µL of high salt buffer (1M K2HPO4, 20mM EDTA, pH 7.8)
- Add 90 µL sample, mix and freeze at -80 °C
- For the measurement thaw by placing the tubes in room temperature water
- Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL-1 lysozyme, 2.5 mg mL-1 BSA, add water for desired volume)
- Mix and incubate the cells for 10 minutes at room temperature
- Prepare the Luciferase Assay Reagent, by adding 10 mL of Luciferase Assay Buffer to the vial containing the Luciferase Assay Substrate
- Fill each well of a white, flat bottom 96 well microtiter plate with 20 µL of cell lysate
- For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a Promega GloMax®-Multi Detection System with dual injector):
- Injection volume of Luciferase Assay Reagent: 100 µL
- Delay: 20 secs
- Integration: 3 secs
References
http://www.promega.com/tbs/tb281/tb281.pdf
User Reviews
UNIQb8d66097d2cc9d8f-partinfo-00000006-QINU UNIQb8d66097d2cc9d8f-partinfo-00000007-QINU