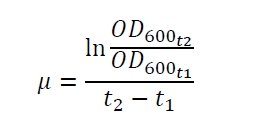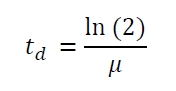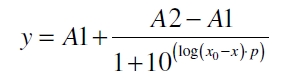Difference between revisions of "Part:BBa K389015:Experience"
(→Data Analysis) |
(→Transfer function) |
||
| (32 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | __TOC__ | |
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
how you used this part and how it worked out. | how you used this part and how it worked out. | ||
===Applications of BBa_K389015=== | ===Applications of BBa_K389015=== | ||
| + | |||
| + | |||
| + | ==== Growth functions and luciferase expression for <partinfo>K389015</partinfo>==== | ||
| + | |||
| + | To characterize this part we performed several cultivations with different concentrations of [http://www.chemblink.com/products/2478-38-8.htm acetosyringone] as inducer and measured the luminescence emitted by the luciferase reaction with luciferin ([[#Protocols|Protocol]]). We used ''Escherichia coli'' DB3.1 carrying the pSB1C3::K389015 plasmid. Even without inducer the bacteria carrying the plasmid showed decelerated growth. In addition acetosyringone affected the growth rates (we used a stock solution of 20 mM acetosyringone solved in 10 % (v/v) DMSO). Growth curves, averaged specific growth rates and doubling times are shown below. It can be observed, that ''E. coli'' carrying the pSB1C3::K389015 plasmid growths nearly linear. | ||
| + | |||
| + | |||
| + | [[Image:K389015growth.jpg|600px|thumb|center|'''Fig. 1: Growth curves for ''E. coli'' DB3.1 without plasmid and carrying <partinfo>K389015</partinfo> with different acetosyringone concentrations in LB medium with 10 mg ml<sup>-1</sup> chloramphenicol.''']] | ||
| + | |||
| + | |||
| + | The specific growth rates µ and doubling times t<sub>d</sub> are calculated with the OD<sub>600</sub> and following formulas: | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_Specific_growth_rate.jpg|175px|center]] <div align="right">(1)</div> | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_Doubling_time.jpg|175px|center]] <div align="right">(2)</div> | ||
| + | |||
| + | |||
| + | <center>Table 1: Averaged specific growth rates and doubling times for cultivations of ''E. coli'' DB3.1 without plasmid and carrying <partinfo>K389015</partinfo> with different acetosyringone concentrations in LB medium with 10 mg ml<sup>-1</sup> chloramphenicol. | ||
| + | |||
| + | {|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | ||
| + | |- | ||
| + | !style="border-style: solid; border-width: 1px"| ''E. coli'' DB3.1 | ||
| + | !style="border-style: solid; border-width: 1px"| µ / h<sup>-1</sup> | ||
| + | !style="border-style: solid; border-width: 1px"| t<sub>d</sub> / h | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| without plasmid | ||
| + | |style="border-style: solid; border-width: 1px"| 0.35 | ||
| + | |style="border-style: solid; border-width: 1px"| 1.98 | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| carrying K389015 | ||
| + | |style="border-style: solid; border-width: 1px"| 0.31 | ||
| + | |style="border-style: solid; border-width: 1px"| 2.24 | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| carrying K389015 with 400 µM acetosyringone | ||
| + | |style="border-style: solid; border-width: 1px"| 0.26 | ||
| + | |style="border-style: solid; border-width: 1px"| 2.67 | ||
| + | |} | ||
| + | |||
| + | </center> | ||
| + | |||
| + | |||
| + | Exemplary induction curves with the luminescence normalized to OD<sub>600</sub> are shown in Fig. 2. We observed a basal transcription, but the induction with acetosyringone is undoubtedly. The detailed [[#Data_Analysis | data analysis]] and [[#Transfer_function | transfer function]] is described below. | ||
| + | |||
| + | |||
| + | [[Image:K389015induction.jpg|600px|thumb|center|'''Fig. 2: Induction curves for ''E. coli'' DB3.1 carrying <partinfo>K389015</partinfo> with different acetosyringone concentrations in LB medium with 10 mg ml<sup>-1</sup> chloramphenicol. The relative luminescence units (RLU) are normalized to OD<sub>600</sub> and plotted against the cultivation time in h''']] | ||
====Transfer function==== | ====Transfer function==== | ||
| − | The data for the transfer function was measured and analyzed as described below. The data was fitted with a | + | The data for the transfer function was measured and analyzed as [[Part:BBa_K389015:Experience#Data Analysis | described below]]. Nelson ''et al.'' suggests using a dose response function and fitting it with a logistical equation for the data analysis of receptor systems ([http://www.nature.com/nature/journal/v416/n6877/abs/nature726.html Nelson ''et al.'', 2002]). The data was fitted with a function of the form |
| − | [[Image:Bielefeld_Doseresponse_fit.jpg|175px|center]] <div align="right">( | + | [[Image:Bielefeld_Doseresponse_fit.jpg|175px|center]] <div align="right">(3)</div> |
| − | with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x<sub>0</sub>). Figure | + | with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x<sub>0</sub>). Figure 3 shows the measured [[Part:BBa_K389015:Experience#Data analysis | ratio ɸ<sub>I</sub>]] between induced (i) and uninduced (u) relative luminescence units (RLU) per OD<sub>600</sub> plotted against the logarithm of the concentration of the inductor [http://www.chemblink.com/products/2478-38-8.htm acetosyringone] in µM. The fit has an R<sup>2</sup> = 0.98. |
| − | [[Image:Bielefeld_Fit_Luc_final.jpg|600px|thumb|center|'''Fig. | + | [[Image:Bielefeld_Fit_Luc_final.jpg|600px|thumb|center|'''Fig. 3: Transfer function for the part <partinfo>K389015</partinfo> (R<sup>2</sup> = 0.98).''']] |
| Line 21: | Line 68: | ||
<center> | <center> | ||
| − | Table | + | Table 2: Data from the transfer function for the part <partinfo>K389015</partinfo>. |
{|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | {|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | ||
| Line 42: | Line 89: | ||
The fully induced VirA/G signaling system with luciferase read-out has a 2.2 fold increased expression compared to the uninduced system. The Hill coefficient is > 1, so a positive cooperation can be observed ([http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 D Chu ''et al.'', 2009]). The [https://parts.igem.org/Switch_Point switch point] of the system is at about 32 µM, so this is the concentration at which the device output is 50% of the maximum output. | The fully induced VirA/G signaling system with luciferase read-out has a 2.2 fold increased expression compared to the uninduced system. The Hill coefficient is > 1, so a positive cooperation can be observed ([http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 D Chu ''et al.'', 2009]). The [https://parts.igem.org/Switch_Point switch point] of the system is at about 32 µM, so this is the concentration at which the device output is 50% of the maximum output. | ||
| − | |||
====Response time==== | ====Response time==== | ||
| Line 48: | Line 94: | ||
The system needs at least one hour to show a measurable reaction to an induction with acetosyringone. In the following illustration the reaction of the system to induction with 200 µM acetosyringone in the exponential growth phase is shown. For a good separation of the induced system from the uninduced system at least two hours are needed. | The system needs at least one hour to show a measurable reaction to an induction with acetosyringone. In the following illustration the reaction of the system to induction with 200 µM acetosyringone in the exponential growth phase is shown. For a good separation of the induced system from the uninduced system at least two hours are needed. | ||
| − | [[Image: | + | [[Image:Bielefeld_Induktionsverhalten_luciferase.jpg|600px|center|thumb|'''Fig. 4: Cultivation of <partinfo>K389015</partinfo> in ''Escherichia coli'' DB3.1. Induction with 200 µM acetosyringone during the exponential growth phase.''']] |
| − | + | ||
====Data Analysis==== | ====Data Analysis==== | ||
| − | Because the luciferase accumulation is very different in different cultivations, the uninduced negative control was used as internal standard. To show the behaviour of the VirA/G signaling system when induced, the ratio ɸ<sub>I</sub> between induced (i) and uninduced (u) relative luminescence | + | Because the luciferase accumulation is very different in different cultivations, the uninduced negative control was used as internal standard. To show the behaviour of the VirA/G signaling system when induced, the ratio ɸ<sub>I</sub> between induced (i) and uninduced (u) relative luminescence units (RLU) per OD<sub>600</sub> is calculated: |
| − | [[Image:Bielefeld_Quotient_RLU.jpg|200px|center]] <div align="right">( | + | [[Image:Bielefeld_Quotient_RLU.jpg|200px|center]] <div align="right">(4)</div> |
| − | As seen above, at least one hour is needed to separate the induced luminescence signal from the uninduced, so ɸ<sub>I</sub> > 1. Within a cultivation ɸ<sub>I</sub> is rising during the first hours and is decreasing after it reached a maximum at OD<sub>600</sub> ~ 1. This is shown in figure 3: | + | [[Part:BBa_K389015:Experience#Response time | As seen above]], at least one hour is needed to separate the induced luminescence signal from the uninduced, so ɸ<sub>I</sub> > 1. Within a cultivation ɸ<sub>I</sub> is rising during the first hours and is decreasing after it reached a maximum at OD<sub>600</sub> ~ 1. This is shown in figure 3: |
| − | [[Image: | + | [[Image:Bielefeld_Luc_fit_ratio_to_OD.jpg|650px|thumb|center|'''Fig. 5A: Typical development of ɸ<sub>I</sub> plotted to OD<sub>600</sub> of a cultivation with <partinfo>K389015</partinfo> in ''E. coli'' DB3.1 with polynomial fit (5th order, R<sup>2</sup> = 0.75, induced with 200 µM acetosyringone). Fig. 5B: Another typical development of ɸ<sub>I</sub> plotted to OD<sub>600</sub> of a cultivation with <partinfo>K389015</partinfo> in ''E. coli'' DB3.1 with polynomial fit (5th order, R<sup>2</sup> = 0.87, induced with 400 µM acetosyringone).''']] |
| − | To measure the ratio of increasing promoter activity by inducing the system ɸ<sub>I</sub> samples for analyzation should be taken at OD<sub>600</sub> | + | To measure the ratio of increasing promoter activity by inducing the system ɸ<sub>I</sub> samples for analyzation should be taken at OD<sub>600</sub> = 1 +/- 0.5. The highest ɸ<sub>I</sub> in this range of the cultivation is taken for the calculation of the [[Part:BBa_K389015:Experience#Transfer function | transfer function]]. |
| + | |||
| + | |||
| + | ====Plasmid conformation analysis==== | ||
| + | A plasmid conformation analysis for the BioBrick <partinfo>K389015</partinfo> in <partinfo>pSB1C3</partinfo> was performed by the [http://web.plasmidfactory.com/de/ PlasmidFactory] by Capillary Gel Electrophoresis (CGE). The chromatogram is shown in fig. 6 and the results in tab. 3. The data shows a high percentage of covalently closed circular (ccc) plasmid DNA. This is the biological active shape of plasmids so a high percentage of ccc plasmid DNA indicates a high quality of plasmid DNA ([http://web.plasmidfactory.com/en/service_CGE.html PlasmidFactory]). | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld_CGE_K389015.jpg|600px|thumb|center|'''Fig. 6: Chromatogram of the CGE of the BioBrick <partinfo>K389015</partinfo> in <partinfo>pSB1C3</partinfo> performed by the [http://web.plasmidfactory.com/de/ PlasmidFactory] (Bielefeld).''']] | ||
| + | |||
| + | |||
| + | <center> | ||
| + | Table 3: Data from the CGE of the BioBrick <partinfo>K389015</partinfo> in <partinfo>pSB1C3</partinfo> performed by the [http://web.plasmidfactory.com/de/ PlasmidFactory] (Bielefeld). | ||
| + | |||
| + | {|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | ||
| + | |- | ||
| + | !style="border-style: solid; border-width: 1px"| Conformation | ||
| + | !style="border-style: solid; border-width: 1px"| Ratio / % | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| ccc monomer | ||
| + | |style="border-style: solid; border-width: 1px"| 91 | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| ccc dimer | ||
| + | |style="border-style: solid; border-width: 1px"| 3.7 | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| oc | ||
| + | |style="border-style: solid; border-width: 1px"| 5.3 | ||
| + | |} | ||
| + | |||
| + | </center> | ||
====Protocols==== | ====Protocols==== | ||
| Line 86: | Line 159: | ||
=====Measurement===== | =====Measurement===== | ||
| − | * For the luciferase detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer | + | * For the [[Part:BBa_K389004 | luciferase]] detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer |
* Prepare reaction tubes with 10 µL of high salt buffer (1M K<sub>2</sub>HPO<sub>4</sub>, 20mM EDTA, pH 7.8) | * Prepare reaction tubes with 10 µL of high salt buffer (1M K<sub>2</sub>HPO<sub>4</sub>, 20mM EDTA, pH 7.8) | ||
| Line 94: | Line 167: | ||
* For the measurement thaw by placing the tubes in room temperature water | * For the measurement thaw by placing the tubes in room temperature water | ||
| − | * Add 300 µL of freshly prepared lysis mix ( | + | * Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL<sup>-1</sup> lysozyme, 2.5 mg mL<sup>-1</sup> BSA, add water for desired volume) |
* Mix and incubate the cells for 10 minutes at room temperature | * Mix and incubate the cells for 10 minutes at room temperature | ||
| Line 104: | Line 177: | ||
* For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a Promega GloMax®-Multi Detection System with dual injector): | * For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a Promega GloMax®-Multi Detection System with dual injector): | ||
** Injection volume of Luciferase Assay Reagent: 100 µL | ** Injection volume of Luciferase Assay Reagent: 100 µL | ||
| − | ** Delay: 20 | + | ** Delay: [[Part:BBa_K389004:Experience#Kinetic of luciferin conversion | 20 s]] |
| − | ** Integration: 3 | + | ** Integration: 3 s |
| − | + | ||
===References=== | ===References=== | ||
Chu D, Zabet NR, Mitavskiy B (2009) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 Models of transcription factor binding: Sensitivity of activation functions to model assumptions], ''J Theor Biol'' 257(3):419-429. | Chu D, Zabet NR, Mitavskiy B (2009) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 Models of transcription factor binding: Sensitivity of activation functions to model assumptions], ''J Theor Biol'' 257(3):419-429. | ||
| + | Greg Nelson, Jayaram Chandrashekar, Mark A. Hoon, Luxin Feng, Grace Zhao, Nicholas J. P. Ryba & Charles S. Zuker (2002) ''[http://www.nature.com/nature/journal/v416/n6877/abs/nature726.html An amino-acid taste receptor ]'', Nature 416: 199-202. | ||
| + | |||
| + | [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System] | ||
| + | |||
| + | [http://web.plasmidfactory.com/en/service_CGE.html PlasmidFactory Homepage] | ||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 00:50, 28 October 2010
Contents
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K389015
Growth functions and luciferase expression for BBa_K389015
To characterize this part we performed several cultivations with different concentrations of [http://www.chemblink.com/products/2478-38-8.htm acetosyringone] as inducer and measured the luminescence emitted by the luciferase reaction with luciferin (Protocol). We used Escherichia coli DB3.1 carrying the pSB1C3::K389015 plasmid. Even without inducer the bacteria carrying the plasmid showed decelerated growth. In addition acetosyringone affected the growth rates (we used a stock solution of 20 mM acetosyringone solved in 10 % (v/v) DMSO). Growth curves, averaged specific growth rates and doubling times are shown below. It can be observed, that E. coli carrying the pSB1C3::K389015 plasmid growths nearly linear.
The specific growth rates µ and doubling times td are calculated with the OD600 and following formulas:
| E. coli DB3.1 | µ / h-1 | td / h |
|---|---|---|
| without plasmid | 0.35 | 1.98 |
| carrying K389015 | 0.31 | 2.24 |
| carrying K389015 with 400 µM acetosyringone | 0.26 | 2.67 |
Exemplary induction curves with the luminescence normalized to OD600 are shown in Fig. 2. We observed a basal transcription, but the induction with acetosyringone is undoubtedly. The detailed data analysis and transfer function is described below.

Transfer function
The data for the transfer function was measured and analyzed as described below. Nelson et al. suggests using a dose response function and fitting it with a logistical equation for the data analysis of receptor systems ([http://www.nature.com/nature/journal/v416/n6877/abs/nature726.html Nelson et al., 2002]). The data was fitted with a function of the form
with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x0). Figure 3 shows the measured ratio ɸI between induced (i) and uninduced (u) relative luminescence units (RLU) per OD600 plotted against the logarithm of the concentration of the inductor [http://www.chemblink.com/products/2478-38-8.htm acetosyringone] in µM. The fit has an R2 = 0.98.
The important data from the transfer function is summarized in table 1:
Table 2: Data from the transfer function for the part BBa_K389015.
| Parameter | Value |
|---|---|
| Hill coefficient | 1.092 |
| Switch point | 31.6 µM |
| Top asymptote | 2.16 |
The fully induced VirA/G signaling system with luciferase read-out has a 2.2 fold increased expression compared to the uninduced system. The Hill coefficient is > 1, so a positive cooperation can be observed ([http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 D Chu et al., 2009]). The switch point of the system is at about 32 µM, so this is the concentration at which the device output is 50% of the maximum output.
Response time
The system needs at least one hour to show a measurable reaction to an induction with acetosyringone. In the following illustration the reaction of the system to induction with 200 µM acetosyringone in the exponential growth phase is shown. For a good separation of the induced system from the uninduced system at least two hours are needed.
Data Analysis
Because the luciferase accumulation is very different in different cultivations, the uninduced negative control was used as internal standard. To show the behaviour of the VirA/G signaling system when induced, the ratio ɸI between induced (i) and uninduced (u) relative luminescence units (RLU) per OD600 is calculated:
As seen above, at least one hour is needed to separate the induced luminescence signal from the uninduced, so ɸI > 1. Within a cultivation ɸI is rising during the first hours and is decreasing after it reached a maximum at OD600 ~ 1. This is shown in figure 3:

To measure the ratio of increasing promoter activity by inducing the system ɸI samples for analyzation should be taken at OD600 = 1 +/- 0.5. The highest ɸI in this range of the cultivation is taken for the calculation of the transfer function.
Plasmid conformation analysis
A plasmid conformation analysis for the BioBrick BBa_K389015 in pSB1C3 was performed by the [http://web.plasmidfactory.com/de/ PlasmidFactory] by Capillary Gel Electrophoresis (CGE). The chromatogram is shown in fig. 6 and the results in tab. 3. The data shows a high percentage of covalently closed circular (ccc) plasmid DNA. This is the biological active shape of plasmids so a high percentage of ccc plasmid DNA indicates a high quality of plasmid DNA ([http://web.plasmidfactory.com/en/service_CGE.html PlasmidFactory]).
Table 3: Data from the CGE of the BioBrick BBa_K389015 in pSB1C3 performed by the [http://web.plasmidfactory.com/de/ PlasmidFactory] (Bielefeld).
| Conformation | Ratio / % |
|---|---|
| ccc monomer | 91 |
| ccc dimer | 3.7 |
| oc | 5.3 |
Protocols
Cultivation
- Inoculate 10 mL LB containing desired Antibiotic with glycerol stock
- Cultivate over night at 37 °C and 175 rpm
- Measure the OD600
- Prepare shake flasks with LB, antibiotic and inductor
- For luciferase measurement at least 10 mL starting volume
- Inoculate the main culture with a starting OD600 of 0.1
- Cultivate at 37 °C and 175 rpm
- Take a sample at least every hour and measure the OD600
Measurement
- For the luciferase detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer
- Prepare reaction tubes with 10 µL of high salt buffer (1M K2HPO4, 20mM EDTA, pH 7.8)
- Add 90 µL sample, mix and freeze at -80 °C
- For the measurement thaw by placing the tubes in room temperature water
- Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL-1 lysozyme, 2.5 mg mL-1 BSA, add water for desired volume)
- Mix and incubate the cells for 10 minutes at room temperature
- Prepare the Luciferase Assay Reagent, by adding 10 mL of Luciferase Assay Buffer to the vial containing the Luciferase Assay Substrate
- Fill each well of a white, flat bottom 96 well microtiter plate with 20 µL of cell lysate
- For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a Promega GloMax®-Multi Detection System with dual injector):
- Injection volume of Luciferase Assay Reagent: 100 µL
- Delay: 20 s
- Integration: 3 s
References
Chu D, Zabet NR, Mitavskiy B (2009) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WMD-4V42JG5-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b6431553217aca1129c5b441f4b78425 Models of transcription factor binding: Sensitivity of activation functions to model assumptions], J Theor Biol 257(3):419-429.
Greg Nelson, Jayaram Chandrashekar, Mark A. Hoon, Luxin Feng, Grace Zhao, Nicholas J. P. Ryba & Charles S. Zuker (2002) [http://www.nature.com/nature/journal/v416/n6877/abs/nature726.html An amino-acid taste receptor ], Nature 416: 199-202.
[http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System]
[http://web.plasmidfactory.com/en/service_CGE.html PlasmidFactory Homepage]
User Reviews
UNIQ3953770681b6ecf7-partinfo-0000000F-QINU UNIQ3953770681b6ecf7-partinfo-00000010-QINU