Difference between revisions of "Part:BBa K346003:Experience"
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ===construction of BBa_K346003=== | ||
| + | |||
| + | [[Image:mbp4.jpg]] | ||
| + | |||
| + | |||
| + | To be specific, the entire coding region of the MBP for standard part was amplified by PCR from full length MerR with two pairs of primers. Two of these primers encoded a three-residue bridge, SSG, which does not occur in MerR and was added to facilitate some flexibility in the loop connecting the two dimerization helix. The two PCR products were digested with EcoR I / BamH I, or BamH I / Pst I and cloned into EcoR I / Pst I -digested pSB1K3 in one step (fig), which was verified by sequencing. Then the RBS is prefixed to MBP. | ||
| + | |||
| + | Based on the same strategy, MBP-His6 was constructed by using two different pairs of primers, which is used for MBP expression test by western blot. The two PCR products were digested with Nde I / BamH I, or BamH I / Xho I and cloned into Nde I / Xho I -digested pET 21a, which contains a region encoding six histines, in one step to construct pET 21a – mbp , which was verified by sequencing. | ||
| + | |||
| + | |||
| + | ==Expression Experiment and Function Test: == | ||
| + | |||
| + | To test the function of this part, both expression experiment and function test is necessary. We have verified the size of the expressed proteins with SDS-page and Western blot. Besides, to test the efficiency of mercury binding, we also carried out the function test with ICP-AES with the mercury gradient from 10^-8M to 10^-6M. | ||
| + | |||
| + | |||
| + | == Results: == | ||
| + | |||
| + | '''Expression of proteins''' | ||
| + | |||
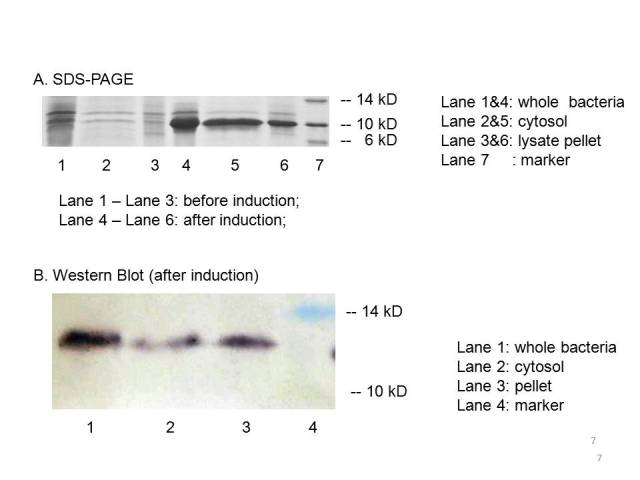
| + | The plasmid PET21a-MBP is transferred to E.coli strain BL21, which can generate T7 polymerase when induced with IPTG. Both induced bacteria and uninduced bacteria(as control) are prepared to make seperate sample of the cytosol, the periplasm and the membrane. The SDS-page and Western blot of the samples showed that compared with the uninduced bacteria, the induced group expressed an protein at the proper place with the size of ~10kD which consists with the predicted size, indicating that the engineered MBP can be expressed in the cytosol. | ||
| + | |||
| + | [[Image:result of MBP.jpg]] | ||
| + | |||
| + | We can detected overexpression band at about 10 kD, which is similar to MBP. Then we fused a his-tag to our target protein and conducted western blot to further verify their expression. Positive band of the expected molecular weight can be detected in the cytosol, which confirm expression and localization of the target protein. We can also indicate from the SDS-PAGE that there exist a large number of MBPs in cytosol which are ready to bind mercury ions. | ||
| + | |||
| + | |||
| + | '''Function test''' | ||
| + | |||
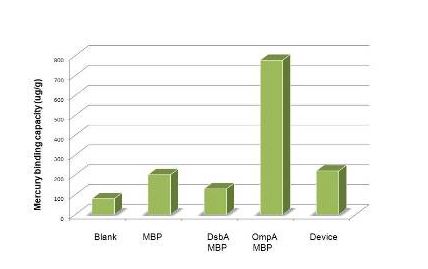
| + | Having made sure that the protein can express normally in the cytosol, the function tests were carried out with ICP-AES. To test the efficiency of mercury absorption of MBP in different concentration of mercury, the concentration gradient from 10^-8M to 10^-6M was set. The results are shown in figure 3. Our experiment shows that the efficiency of MBP increases with the the mercury concentration. Additionally, MBP can bind Hg(II) with high efficiency and sensitivity from the concentration of 10^-7 M(not show in the figure). | ||
| + | |||
| + | [[Image:dfadsf v2.jpg]] | ||
| + | |||
| + | Figure 3: Overnight culture was diluted 1:100 and cultured until OD600=0.6-1.0. We then resuspended the culture with 1% PBS after the centrifugation. Samples for ICP-AES were made following the protocol(See more about this, please visit our wiki.) | ||
| − | |||
| − | |||
| − | |||
| − | |||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 04:46, 28 October 2010
Contents
construction of BBa_K346003
To be specific, the entire coding region of the MBP for standard part was amplified by PCR from full length MerR with two pairs of primers. Two of these primers encoded a three-residue bridge, SSG, which does not occur in MerR and was added to facilitate some flexibility in the loop connecting the two dimerization helix. The two PCR products were digested with EcoR I / BamH I, or BamH I / Pst I and cloned into EcoR I / Pst I -digested pSB1K3 in one step (fig), which was verified by sequencing. Then the RBS is prefixed to MBP.
Based on the same strategy, MBP-His6 was constructed by using two different pairs of primers, which is used for MBP expression test by western blot. The two PCR products were digested with Nde I / BamH I, or BamH I / Xho I and cloned into Nde I / Xho I -digested pET 21a, which contains a region encoding six histines, in one step to construct pET 21a – mbp , which was verified by sequencing.
Expression Experiment and Function Test:
To test the function of this part, both expression experiment and function test is necessary. We have verified the size of the expressed proteins with SDS-page and Western blot. Besides, to test the efficiency of mercury binding, we also carried out the function test with ICP-AES with the mercury gradient from 10^-8M to 10^-6M.
Results:
Expression of proteins
The plasmid PET21a-MBP is transferred to E.coli strain BL21, which can generate T7 polymerase when induced with IPTG. Both induced bacteria and uninduced bacteria(as control) are prepared to make seperate sample of the cytosol, the periplasm and the membrane. The SDS-page and Western blot of the samples showed that compared with the uninduced bacteria, the induced group expressed an protein at the proper place with the size of ~10kD which consists with the predicted size, indicating that the engineered MBP can be expressed in the cytosol.
We can detected overexpression band at about 10 kD, which is similar to MBP. Then we fused a his-tag to our target protein and conducted western blot to further verify their expression. Positive band of the expected molecular weight can be detected in the cytosol, which confirm expression and localization of the target protein. We can also indicate from the SDS-PAGE that there exist a large number of MBPs in cytosol which are ready to bind mercury ions.
Function test
Having made sure that the protein can express normally in the cytosol, the function tests were carried out with ICP-AES. To test the efficiency of mercury absorption of MBP in different concentration of mercury, the concentration gradient from 10^-8M to 10^-6M was set. The results are shown in figure 3. Our experiment shows that the efficiency of MBP increases with the the mercury concentration. Additionally, MBP can bind Hg(II) with high efficiency and sensitivity from the concentration of 10^-7 M(not show in the figure).
Figure 3: Overnight culture was diluted 1:100 and cultured until OD600=0.6-1.0. We then resuspended the culture with 1% PBS after the centrifugation. Samples for ICP-AES were made following the protocol(See more about this, please visit our wiki.)
User Reviews
UNIQ0edc646408313428-partinfo-00000000-QINU UNIQ0edc646408313428-partinfo-00000001-QINU