Difference between revisions of "Part:BBa J45992"
Mkakadiari (Talk | contribs) (→B0030 vs B0034 in E.coli BL21 (DE3) - Thessaly 2024) |
Mkakadiari (Talk | contribs) (→B0030 vs B0034 in E.coli BL21 (DE3) - Thessaly 2024) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 43: | Line 43: | ||
<b> Methods </b> <br> | <b> Methods </b> <br> | ||
| − | Measurements for OD 600nm and fluorescence (488nm, | + | Measurements for OD 600nm and fluorescence (488nm,515nm) were taken over the course of 16 hours, in a 96-well microplate. Clonings were done according to the Golden Braid method, leaving us with level α constructs in the pDGB3a1 backbone (<html> <a href="https://parts.igem.org/Part:BBa_K4213058">BBa_K4213058. </a> </html>) <br> |
| − | + | Constructs and pDGB3a1 (empty vector) transformed into <i> E.coli </i>BL21 (DE3) chassis, incubated at 37oC, 180rpm for 16 hours. <br> | |
| − | BBa_J45992 was inserted in the constructs <html> <a href="https://parts.igem.org/Part:BBa_K5299204">BBa_K5299204,</a><a href="https://parts.igem.org/Part:BBa_K5299205">BBa_K5299205.</a></html> <br> | + | BBa_J45992 was inserted in the constructs <html> <a href="https://parts.igem.org/Part:BBa_K5299204">BBa_K5299204,</a><a href="https://parts.igem.org/Part:BBa_K5299205">BBa_K5299205.</a></html> For more information, head over to these pages. <br> |
| − | Plated | + | Plated 200 ul 5 times, out of each single liquid bacterial culture, created from the same bacterial colony, in order to establish accuracy through technical repeats. <br> |
Medium used was M9 due to minimal interference, with D-glucose serving as the carbon source. Also, served as blank, plated 5 times. <br> | Medium used was M9 due to minimal interference, with D-glucose serving as the carbon source. Also, served as blank, plated 5 times. <br> | ||
| Line 56: | Line 56: | ||
<b> Results </b> <br> | <b> Results </b> <br> | ||
| − | Measurements were taken over the course of 16 hours. Here are the results for BBa_J45992 regarding the different constructs created. <br> | + | Measurements were taken over the course of 16 hours. Here are the results for BBa_J45992 regarding the different constructs created with a pDGB3a1 backbone.. <br> |
<html> | <html> | ||
<center> | <center> | ||
<figure> | <figure> | ||
| − | <img src='https://static.igem.wiki/teams/5299/mar/j45992-b0030-vs-b0034.png' width='700px' height='435px' | + | <img src='https://static.igem.wiki/teams/5299/mar/new-j45992-b0030-vs-b0034.png' width='700px' height='435px' |
| − | <thumb><center><b><small><i>Figure 1: Constructs with the | + | <thumb><center><b><small><i>Figure 1: Constructs with the BBa_J45992 promoter, the BBa_B0030 or the BBa_B0034 RBS, the BBa_I746916 sfGFP and the BBa_B0015 terminator. The pDGB3a1 backbone served as a negative control.</i></small></b></center></thumb> |
</figure></center></html><br> | </figure></center></html><br> | ||
| − | + | ||
| − | + | ||
Latest revision as of 21:09, 30 September 2024
Full-length stationary phase osmY promoter
BBa_J45992 is a stationary phase promoter derived from the promoter that controls transcription of osmY in Escherichia coli Yim-1992, Yim-1994. BBa_J45992 is active in stationary phase and under high osmotic pressure conditions.
Usage and Biology
Expression of osmY is dependent on σS in vivo Hengge-Aronis-1993. Schellhorn et al. previously demonstrated that an osmY::lacZ fusion generated the highest transcriptional signal in stationary phase as compared to nine other σS-dependent promoter-lacZ fusions Schellhorn-1998, Vijayakumar-2004. In addition, the osmY::lacZ fusion generated only a small transcriptional signal during exponential growth.
Contribution: Macquarie University 2019
Group: Macquarie_Australia
Authors: Samuel Beach
Summary: We compared the expression of our cyclic-di-GMP riboswitch + eGFP bioparts over 60 hours of incubation between this, a Lac (BBa_R0010), and a Tac [BBa_K180000] promoter. All three promoters showed significant increase in expression starting at the stationary phase, however the Lac and Tac promoters showed a linear increase from that point while the Full-length stationary phase osmY promoter significantly increased expression then plateued after several hours.
In order to characterise the mode of action of the riboswitch/promoter combinations, we collaborated with Team Sydney Australia to perform GFP fluorescence assays[1]. These assays were used to quantify the production of eGFP between our reporter constructs, as well as comparing the function of the stationary phase promoter against two inducible promoters, Lac [BBa_R0010] and Tac [BBa_K180000]. Samples were measured with the BMG Pherastar plate reader to measure eGFP (Ex485 nm Em520 nm) and OD (600 nm).
These results demonstrate that while the production of eGFP increases at 6-8 hours for all the promoters (i.e. roughly the point at which samples would be entering the stationary phase), the Lac (Fig. 1 & 4) and Tac (Fig. 2 & 5) inducible promoters show a steady, somewhat linear accumulation of eGFP over time, the stationary phase promoter (Fig. 3) shows a sharp increase and beings to level out after ~20 hours.
These results are consistent with previous characterisations of the promoter, which observed significant increases in β-galactosidase concentration after entry into the stationary phase at 5 hours[2].
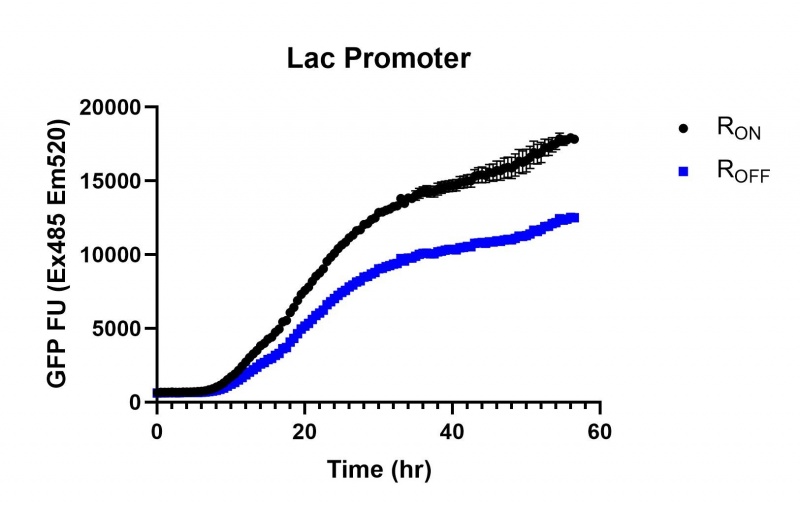
Figure 1: eGFP produced by cells transformed with the Lac riboswitch constructs [BBa_K3151028] over 60 hours.
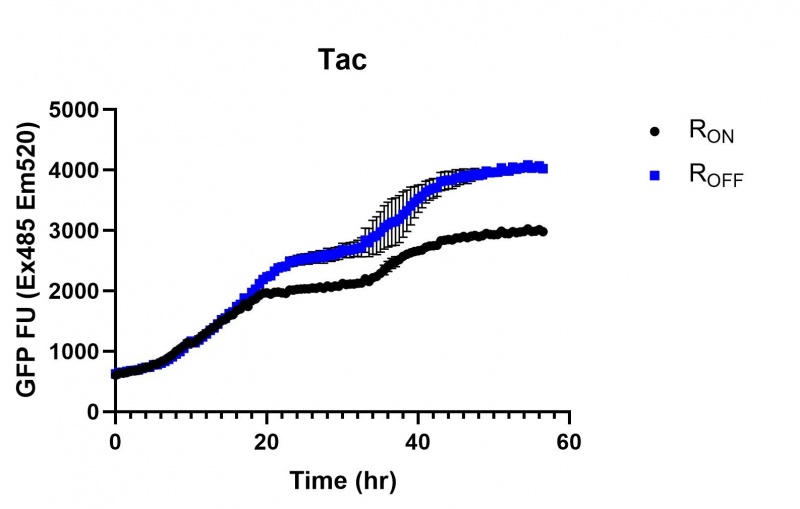
Figure 2: eGFP produced by cells transformed with the Tac riboswitch constructs [BBa_K180000] over 60 hours. We believe that the reason the Tac promoter has the opposite action of the other promoters is due to the fact that the binding of cyclic-di-GMP to the riboswitch results in shorter transcripts, allowing the RNA polymerase to produce more of the transcripts quickly and saturating the cyclic-di-GMP, resulting in a loss of inhibition of eGFP transcription.
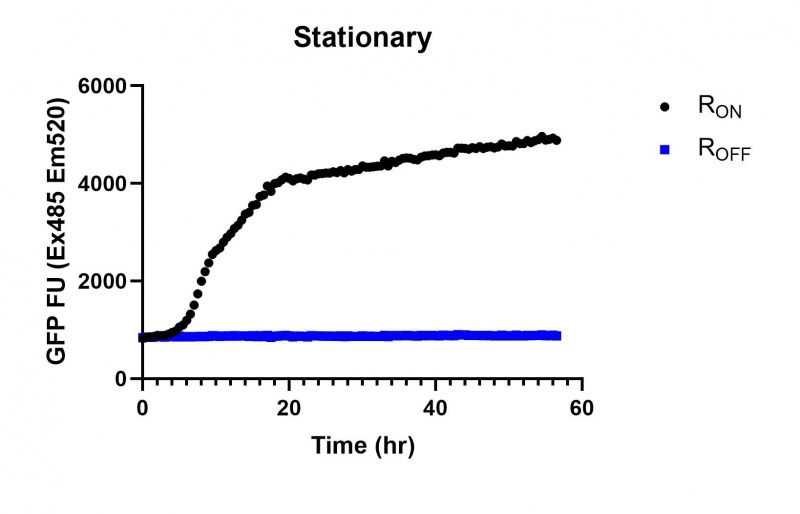
Figure 3: eGFP produced by cells transformed with the Stationary phase riboswitch constructs [BBa_K3151011] over 60 hours.
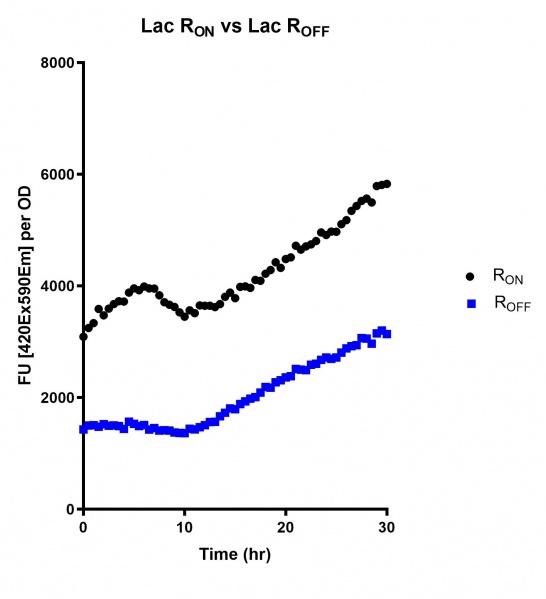
Figure 4: eGFP produced by cells transformed with the Lac riboswitch constructs [BBa_K3151028] over 30 hours. Data provided by Usyd iGEM.
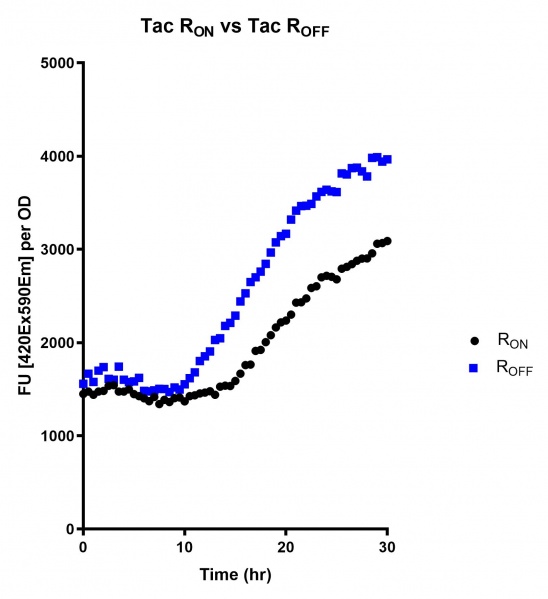
Figure 5: eGFP produced by cells transformed with the Tac riboswitch constructs [BBa_K180000] over 30 hours. Data provided by Usyd iGEM. We believe that the reason the Tac promoter has the opposite action of the other promoters is due to the fact that the binding of cyclic-di-GMP to the riboswitch results in shorter transcripts, allowing the RNA polymerase to produce more of the transcripts quickly and saturating the cyclic-di-GMP, resulting in a loss of inhibition of eGFP transcription.
B0030 vs B0034 in E.coli BL21 (DE3) - Thessaly 2024
Group: Thessaly 2024
Purpose
BBa_J45992 was used in our promoter testing experiments as a positive control for phase activation. It was expected to be working efficiently during the stationary phase of the bacterial population’s life cycle, according to already existing information in the Registry.
Methods
Measurements for OD 600nm and fluorescence (488nm,515nm) were taken over the course of 16 hours, in a 96-well microplate. Clonings were done according to the Golden Braid method, leaving us with level α constructs in the pDGB3a1 backbone ( BBa_K4213058. )
Constructs and pDGB3a1 (empty vector) transformed into E.coli BL21 (DE3) chassis, incubated at 37oC, 180rpm for 16 hours.
BBa_J45992 was inserted in the constructs BBa_K5299204,BBa_K5299205. For more information, head over to these pages.
Plated 200 ul 5 times, out of each single liquid bacterial culture, created from the same bacterial colony, in order to establish accuracy through technical repeats.
Medium used was M9 due to minimal interference, with D-glucose serving as the carbon source. Also, served as blank, plated 5 times.
Measurements were normalised as such: using the average price of fluorescence for the 5 technical repeats and dividing it by the average price of OD.
Standard deviation included in the graphs.
Results
Measurements were taken over the course of 16 hours. Here are the results for BBa_J45992 regarding the different constructs created with a pDGB3a1 backbone..

References:
- Dell EJ. Bottom reading of cell-based assays: direct optic approach enhances fluorescent protein and other microplate analyses. Genetic Engineering & Biotechnology News. 2012 May 1;32(9):22-3.
- Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. Journal of Bacteriology. 1993 Jan 1;175(1):259-65.
<biblio>
- Yim-1994 pmid=8282684
- Yim-1992 pmid=1317380
- Hengge-Aronis-1993 pmid=8416901
- Vijaykumar-2004 pmid=15576800
- Schellhorn-1998 pmid=9829938
</biblio>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]