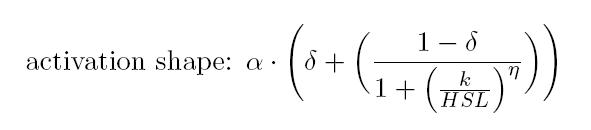Difference between revisions of "Part:BBa T9002:Experience"
(→User Reviews) |
|||
| (34 intermediate revisions by 8 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | + | This experience page is provided so that any user may enter their experience using this part.<BR> |
| − | how you used this part and how it worked out. | + | Please enter how you used this part and how it worked out. |
| − | + | ||
| − | + | ||
===User Reviews=== | ===User Reviews=== | ||
| − | <!-- DON'T DELETE --><partinfo> | + | <!-- DON'T DELETE --><partinfo>BBa_K1157006 StartReviews</partinfo> |
<!-- Template for a user review | <!-- Template for a user review | ||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
| Line 17: | Line 15: | ||
|}; | |}; | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
| + | |||
| + | |||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%' valign='top'| | ||
| + | <partinfo>BBa_T9002 AddReview 1</partinfo> | ||
| + | <I>iGEM_Sevilla_2011</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | <html><a name='t9002PV11'></a></html> | ||
| + | |||
| + | We used this construction as a model for a GFP-based characterization | ||
| + | |||
| + | method. We tried different concentrations, that turned to be too high, | ||
| + | |||
| + | for it seems that the media was quite saturated. | ||
| + | |||
| + | <nowiki>Fluorescence | ||
| + | |||
| + | ---------------------------------------------------------- O.D. | ||
| + | |||
| + | (600nm)</nowiki> | ||
| + | |||
| + | [[Image:Fluabs.png]] | ||
| + | |||
| + | |||
| + | Fluorescence/Absorbance ratio and average of these data | ||
| + | |||
| + | [[Image:Fluoabsavrg.png]] | ||
| + | |||
| + | |||
| + | Variation of the fluo/Abs ratio in time | ||
| + | |||
| + | [[Image:Variationfluoabs.png]] | ||
| + | |||
| + | |||
| + | [[Image:GFPgraph.png]] | ||
| + | |||
| + | |||
| + | |||
| + | <!-- DON'T DELETE --><partinfo>BBa_T9002 EndReviews</partinfo> | ||
| + | |}; | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%' valign='top'| | ||
| + | <partinfo>BBa_T9002 AddReview 2</partinfo> | ||
| + | <I>iGEM_Pavia_2011</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | ''Antiquity. This review comes from the old result system and indicates that this part worked in some test.'' | ||
| + | |||
'''2009 DNA Distribution quality control''' | '''2009 DNA Distribution quality control''' | ||
| − | The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was completely confirmed, while iGEM QC results classified it as "inconsistent". DNA was resuspended from well 9A, kit plate 2, transformed in TOP10 E. coli and amplified inoculating a single colony from the grown LB agar plate in LB medium. Finally DNA has been miniprepped from the grown culture and sent to a BMR Genomics (Padova, Italy) for sequencing. | + | The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was |
| + | |||
| + | completely confirmed, while iGEM QC results classified it as | ||
| + | |||
| + | "inconsistent". DNA was resuspended from well 9A, kit plate 2, | ||
| + | |||
| + | transformed in TOP10 E. coli and amplified inoculating a single colony | ||
| + | |||
| + | from the grown LB agar plate in LB medium. Finally DNA has been | ||
| + | |||
| + | miniprepped from the grown culture and sent to a BMR Genomics (Padova, | ||
| + | |||
| + | Italy) for sequencing. | ||
'''Experimental measurements''' | '''Experimental measurements''' | ||
| − | |||
| − | The | + | The UNIPV-Pavia iGEM team tested T9002 BioBrick in several working |
| − | + | conditions. Results are reported in <partinfo>BBa_F2620</partinfo> | |
| − | + | Experience page. | |
| + | The Brown iGEM team conducted tests on this part in the summer of 2007. | ||
| + | |||
| + | The results are depicted in the graphs below. | ||
| + | |||
| + | [[Image:AHL.JPG|700px]] | ||
| + | |||
| + | The first graph indicates that until a critical point is reached, | ||
| + | |||
| + | increasing AHL concentration does increase GFP output. This is most | ||
| + | |||
| + | easily noticeable between the concentrations of 20 nM, after which | ||
| + | |||
| + | point the amount of GFP produced by cells begins to decrease. This may | ||
| + | |||
| + | be due to one of two things: AHL quenches the signal from GFP, or too | ||
| + | |||
| + | much AHL disrupts the cell's functions in a way that either kills it or | ||
| + | |||
| + | prevents it from making as much GFP. This second hypothesis is | ||
| + | |||
| + | partially confirmed by the second graph, which shows that adding more | ||
| + | |||
| + | than 20 nM AHL causes a decline in cell density. On each graph, the | ||
| + | |||
| + | different colored lines represent different time points after AHL was | ||
| + | |||
| + | added to the cells. They are 4 hours, 5 hours, etc. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | <html> | ||
| + | |||
| + | BioBrick <a href="https://parts.igem.org/Part:BBa_T9002">BBa_T9002</a> | ||
| + | |||
| + | is an HSL biosensor, which provides a non linear relationship between | ||
| + | |||
| + | HSL input and S<sub>cell</sub> output. More precisely, the | ||
| + | |||
| + | characteristic sigmoidal curve requires synthetic parameters for its | ||
| + | |||
| + | accurate identification. These are the minimum and maximum values, the | ||
| + | |||
| + | swtich point (i.e., the curve inflection point), and the upper and | ||
| + | |||
| + | lower boundaries of linearity. This biosensor revealed greatly | ||
| + | |||
| + | reliable, providing measurement repeatability and minimal experimental | ||
| + | |||
| + | noise. Referring to its activation formula, the calibration curve is | ||
| + | |||
| + | shown below.<br></div> | ||
| + | <br> | ||
| + | <div style='text-align:center'> | ||
| + | <div class="thumbinner" style="width:100%;"> <a | ||
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg"><img | ||
| + | |||
| + | alt="" src="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg" | ||
| + | |||
| + | class="thumbimage" width="47%" height="50%"></a></div> | ||
| + | </div> | ||
| + | <br> | ||
| + | <div style='text-align:center'> | ||
| + | <div class="thumbinner" style="width:100%;"> <a | ||
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg"><img | ||
| + | |||
| + | alt="" src="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg" | ||
| + | |||
| + | class="thumbimage" width="100%"></a></div> | ||
| + | </div> | ||
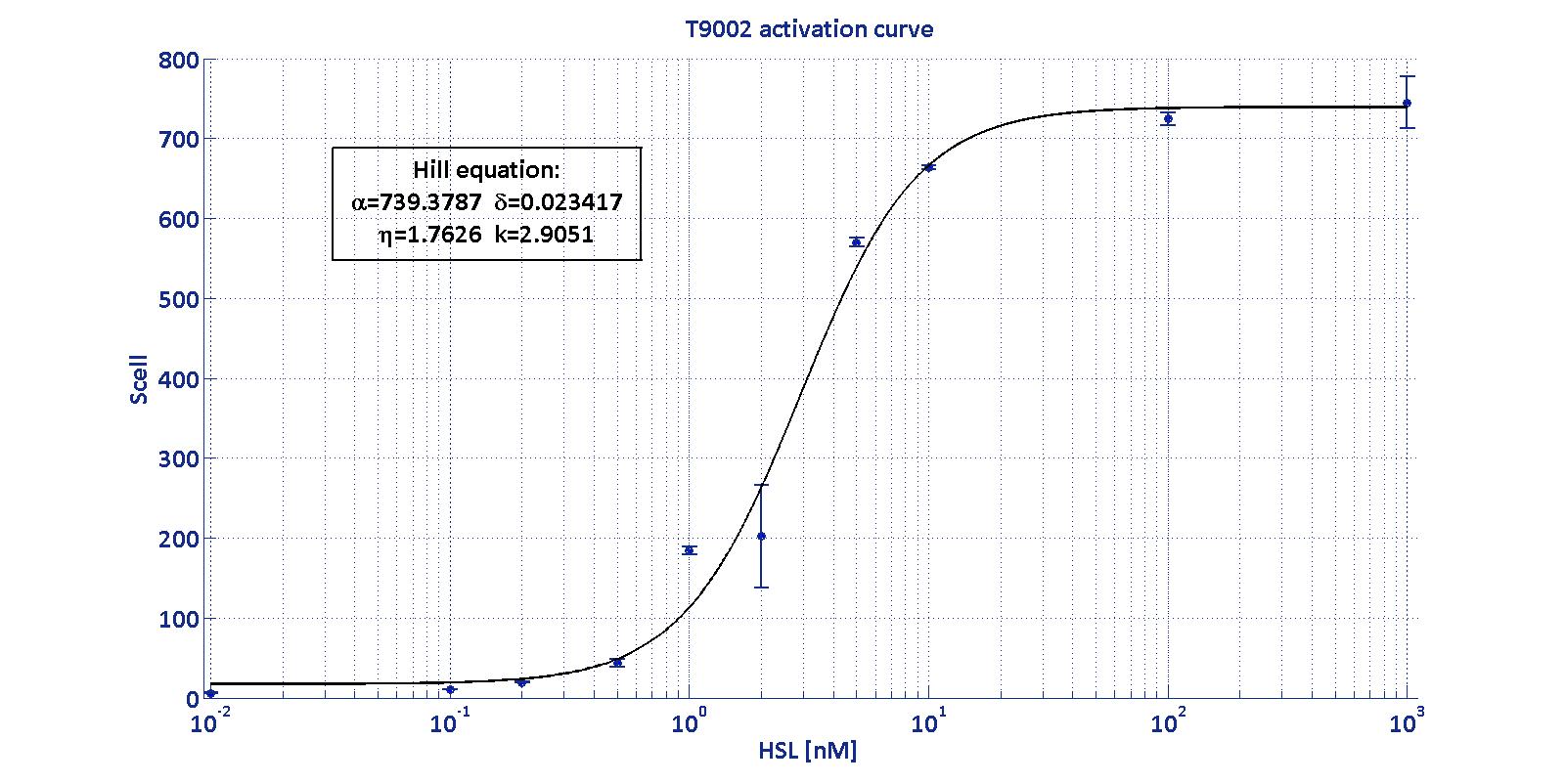
| + | <center> | ||
| + | <table width='50%' class='data' border='1'> | ||
| + | <tr> | ||
| + | <td class='row'><b>Minimum [S<sub>cell</sub>]</b></td> | ||
| + | <td class='row'><b>Maximum [S<sub>cell</sub>]</b></td> | ||
| + | <td class='row'><b>Switch point [nM]</b></td> | ||
| + | <td class='row'><b>Lower boundary of linearity [nM]</b></td> | ||
| + | <td class='row'><b>Upper boundary of linearity [nM]</b></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class='row'>17.31</td> | ||
| + | <td class='row'>739.4</td> | ||
| + | <td class='row'>1.39</td> | ||
| + | <td class='row'>0.38</td> | ||
| + | <td class='row'>5.07</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div align="justify">In order to determine the threshold sensitivity of | ||
| + | |||
| + | T9002 biosensor, experiments were performed with several HSL inductions | ||
| + | |||
| + | minimally interspaced in the region of low detectability. Hypothesizing | ||
| + | |||
| + | that the inducer is 1:20 diluted (as for all of our tests), the minimum | ||
| + | |||
| + | detectable HSL concentration is 3 nM.</div> | ||
| + | <br><br> | ||
| + | <a name='t9002PV11'></a> | ||
| + | This biosensor was used to measure HSL concentration for parts | ||
| + | |||
| + | producing or degrading this signalling molecule. For each of these | ||
| + | |||
| + | experiments, a calibration curve of the biosensor was built, inducing | ||
| + | |||
| + | it with known HSL concentrations, evaluating for each of them the | ||
| + | |||
| + | S<sub>cell</sub> signal and finally estimating the Hill curve | ||
| + | |||
| + | parameters. Once identified the parameters, the unknown concentration | ||
| + | |||
| + | of a sample can be evaluated from its S<sub>cell</sub> (provided that | ||
| + | |||
| + | it has a value included in the linear zone of the biosensor), as shown | ||
| + | |||
| + | below: | ||
| + | |||
| + | <div class="center"><div class="thumbinner" style="width: 650px;"><a | ||
| + | |||
| + | href="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG" | ||
| + | |||
| + | class="image"><img alt="" | ||
| + | |||
| + | src="https://static.igem.org/mediawiki/2011/e/e2/UNIPV_HSL.PNG"class="thumbim | ||
| + | |||
| + | age" width="40%"></a></div></div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | It is necessary, then, to multiply the measurement for the dilution | ||
| + | |||
| + | factor used (in our experiments it was 20). | ||
| + | |||
| + | </html> | ||
| + | |}; | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%' valign='top'| | ||
| + | <partinfo>BBa_T9002 AddReview 3</partinfo> | ||
| + | <I>iGEM_Aberdeen_2014</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | ==== Team Aberdeen iGEM 2014 Characterisation of BioBrick T9002 ==== | ||
| + | |||
| + | This BioBrick is a GFP Expresser Controlled by 3OC6HSL Receiver Device. | ||
| + | |||
| + | BioBrick BBa_T9002 is a composite part encoding a quorum sensing (QS) receiver driving expression of green fluorescent protein. | ||
| + | |||
| + | |||
| + | ==== Aims and Rationale ==== | ||
| + | T9002 was previously reported to exhibit increased GFP expression in the presence of AHL (N-acyl homoserine lactone). | ||
| + | |||
| + | We aimed to confirm GFP-expression responsiveness and dependence of AHL, and how the physical proximity of this QS receiver to any QS sender (producer of homoserine lactone) is facilitated by surface-binding affects quorum signalling. | ||
| + | |||
| + | ==== Materials and Methods ==== | ||
| + | |||
| + | T9002 on a pSB1C3 backbone was rescued from Spring 2014 Distribution Plate 3-22J and transformed into XL1Blue ''E. coli'' competent cells, and selected on Chloramphenicol LB agar. | ||
| + | |||
| + | Chloramphenicol LB liquid was innoculated with T9002 ''E. coli'' transformant and these were grown in conjunction with other ''E. coli'' transformed with the AHL ‘Sender’ plasmid [[Part:BBa_K1090000]] (also on pSB1C3). BBa_K1090000 codes for AHL-synthesising enzymes, and also constitutively expresses RFP. | ||
| + | |||
| + | ===== Poly-L-lysine Cell Adhesion ===== | ||
| + | To investigate QS signalling, and how it is affected by binding of sender and receiver bacteria to a solid surface, we immobilised our cells on a surface coated with poly-lysine: | ||
| + | |||
| + | # 50µl 0.01% Poly-Lysine was added to each well of a 96 well glass-bottom Plate (Whatman), and incubated at room temperature for 2 hours | ||
| + | # Excess poly-lysine was removed and the plate washed three times with sterile water and allowed to dry at room temperature. | ||
| + | # E. coli cultures were grown overnight in a 37C shaking water bath, and diluted to an optical density-600nm(OD600) of 0.02. | ||
| + | # K1090000 ‘Sender’ transformants washed by centrifugation at 13000rpm for 1minute and resuspension in phosphate buffered saline (PBS), three times. | ||
| + | # Dual Sender-Receiver wells were diluted and combined in a colorimetric ratio to achieve a total OD600 of 0.02, with a range of 1:1 to 1:10,000,000 Sender:Receiver. 50µl total volume in Liquid Broth (LB) used per well. Incubated at 4C for 1 hour. | ||
| + | # Unbound cells removed by lightly shaking over waste and washed three times with Phosphate-Buffered Saline (PBS). 50µl fresh LB medium was added to each well. | ||
| + | # A FluoSTAR OPTIMA Fluoresence Plate Reader was used to measure Red(Excitation 544nm/Emission 612nm) and Green(Excitation 485nm/Emission 520nm) Fluorescence was recorded every 5 minutes for 7 hours; this incubates the samples at 37 degrees C and shakes (1mm double-orbital) for 30 seconds before each read. | ||
| + | |||
| + | ==== Results ==== | ||
| + | K1090000 RFP expression RFP expression of K1090000 transformed E. coli was compared against untransformed XL1-Blue E. coli negative control and an RFP pSB1C3 plasmid positive control. Analysis of red fluorescence using a fluorimeter plate reader clearly shows that K1090000 constitutively expressed moderate amounts of RFP (Figure 1). | ||
| + | |||
| + | ===== T9002 with AHL ===== | ||
| + | T9002 E. coli was resuspended in conditioned growth medium containing AHL, derived from a filter sterilised K1090000 suspension. The T9002 transformants in this medium as expected displayed a slow, exponential increase in GFP, indicating the T9002 receiver cells were responding to the QS signalling caused by AHL in the medium. However, QS responses by the T9002 receivers were even more marked when T9002 transformants were co-cultured with K109000 QS sender transformants (Figure 2). | ||
| + | |||
| + | [[File:FIG1-T9002.png|600px]] | ||
| + | |||
| + | ''Figure 1 - K1090000 E. coli constitutively expresses RFP. Red (Excitation 544nm/Emission 612nm) fluorescence of K1090000 E. coli (triangles), RFP positive control pSB1C3 (Circles), untransformed XL1-Blue E. coli (dashes).'' | ||
| + | |||
| + | |||
| + | T9002 with K1090000 We observed that optimal conditions for absolute green fluorescent production by T9002 receiver transformants were a ‘sender’ (S) to ‘receiver’ (R) ratio of 1:100; Figure 3. It was inferred that ratios greater or smaller than this resulted in too little AHL or undesirable competition effects, preventing optimal GFP fluorescence. An untransformed XL1-Blue E. coli (X) acted as control. | ||
| + | T9002 Receivers in the presence of K1090000 Sender exhibit significantly higher GFP response than T9002 with a control (non-AHL expressing untransformed XL1-Blue E. coli); Figure 4. | ||
| + | |||
| + | |||
| + | [[File:FIG2-T9002.png|600px]] | ||
| + | |||
| + | ''Figure 2 – Production of GFP by T9002, induced either by filtered AHL-containing culture medium, or by actively growing K1090000 sender transformants. AHL derived from culture medium is sufficient for slow rate T9002 Receiver GFP production (-), although T9002 GFP production was more efficient when paired with actively-growing K1090000 senders (circles). Mean GFP fluorescence of E.coli free in 50µl suspensions incubated at 37C, 1mm double-orbital shaking for 30 seconds every 5 minutes for 355 minutes.'' | ||
| + | |||
| + | [[File:FIG3-T9002(3).png|600px]] | ||
| + | |||
| + | ''Figure 3 – Greatest absolute fluorescence by T9002 transformants is observed in a Sender (S) 1:100 Receiver (R) cell number initial ratio. T9002 Receiver-produced GFP was compared with that produced when paired with an untransformed XL1-Blue E. coli (X) control or K1090000 Senders (circles), in triplicate. 50µl suspensions were incubated at 37C, 1mm double-orbital shaking for 30 seconds every 5 minutes for 355 minutes.'' | ||
| + | |||
| + | [[File:FIG4-T9002.png|600px]] | ||
| + | |||
| + | ''Figure 4 – K1090000 Sender E. coli induction increased GFP expression in T9002 Receivers. Growth started in cell number ratios of aliquots totalling OD600 0.02 densities, suspended in liquid LB. Untransformed XL1-Blue:Receiver 1:100 ratio (Filled Diamonds), Sender:Receiver 1:100 ratio (Open circles). Values are blank corrected.'' | ||
| + | |||
| + | '''T9002 and K1090000 bound to a Poly-Lysine surface.''' Overall, immobilised Sender/Receiver pairs exhibited a greater extended rate of GFP production response, and higher absolute response after 7 hours than sender-receiver pairs free in suspension; Figure 5. | ||
| + | |||
| + | [[File:FIG5-T9002.png|600px]] | ||
| + | |||
| + | ''Figure 5: Surface-immobilisation of sender-receiver QS tranformants results in improved GFP expression. Poly-L-lysine wells (triangles) had greater rate of response compared to cells free in suspension (circles). This was performed in a Sender 1:10 Receiver cell number ratio.'' | ||
| + | |||
| + | |||
| + | ==== Conclusions ==== | ||
| + | |||
| + | * BBa_T9002 is an AHL receiver coupled to a GFP reporter, it has ‘leaky’ GFP expression. | ||
| + | * BBa_T9002 GFP production significantly increases in the presence of AHL. | ||
| + | * BBa_K1090000 ‘Sender’ is an effective AHL expressor that is able to activate BBa_T9002 ‘Receiver’ GFP production. | ||
| + | * BBa_K1090000 has moderate constitutive RFP expression. | ||
| + | * Poly-Lysine surface-immobilisation of T9002 and K1090000 ''E. coli'' extends the longterm production rate of GFP expression. | ||
| + | |}; | ||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | <partinfo>BBa_T9002 AddReview | + | <partinfo>BBa_T9002 AddReview 4</partinfo> |
| − | <I> | + | <I>iGEM_Stockholm_2015</I> |
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | + | ||
| − | + | '''Method:''' For each experiment fresh solutions in was prepared in ddH2O in the following concentrations: 100 mM; 10 mM; 0.1 mM; 0.01 mM. We tested these different concentrations in 3 repetitions first in agar plates with the bacteria with BBa_T9002. Then we tested these different concentrations in the liquid LB cultures with 3 repetitions for each sample. Both plates and liquid cultures were incubated in 37°C for 24h. | |
| − | + | ||
| + | '''Results:''' No color change was observed for any of the experiments. It seems that our bacterial strain did not respond to any concentrations of OHHL in agar or liquid cultures. | ||
| + | |||
| + | '''Conclusion:''' Based on our results, this part of our experiment did not work as intended. | ||
| + | |||
| + | Please, visit our [http://2015.igem.org/Team:Stockholm iGEM Stockholm 2015 webpage] for more details or contact us. | ||
| + | |}; | ||
Latest revision as of 16:14, 18 September 2015
This experience page is provided so that any user may enter their experience using this part.
Please enter how you used this part and how it worked out.
User Reviews
UNIQa8a54d2bec99b200-partinfo-00000000-QINU
|
•
iGEM_Sevilla_2011 |
We used this construction as a model for a GFP-based characterization method. We tried different concentrations, that turned to be too high, for it seems that the media was quite saturated. Fluorescence ---------------------------------------------------------- O.D. (600nm)
UNIQa8a54d2bec99b200-partinfo-00000004-QINU |
|
••
iGEM_Pavia_2011 |
Antiquity. This review comes from the old result system and indicates that this part worked in some test.
The UNIPV-Pavia iGEM team sequenced T9002 part and found that it was completely confirmed, while iGEM QC results classified it as "inconsistent". DNA was resuspended from well 9A, kit plate 2, transformed in TOP10 E. coli and amplified inoculating a single colony from the grown LB agar plate in LB medium. Finally DNA has been miniprepped from the grown culture and sent to a BMR Genomics (Padova, Italy) for sequencing. Experimental measurements The UNIPV-Pavia iGEM team tested T9002 BioBrick in several working conditions. Results are reported in BBa_F2620 Experience page. The Brown iGEM team conducted tests on this part in the summer of 2007. The results are depicted in the graphs below. The first graph indicates that until a critical point is reached, increasing AHL concentration does increase GFP output. This is most easily noticeable between the concentrations of 20 nM, after which point the amount of GFP produced by cells begins to decrease. This may be due to one of two things: AHL quenches the signal from GFP, or too much AHL disrupts the cell's functions in a way that either kills it or prevents it from making as much GFP. This second hypothesis is partially confirmed by the second graph, which shows that adding more than 20 nM AHL causes a decline in cell density. On each graph, the different colored lines represent different time points after AHL was added to the cells. They are 4 hours, 5 hours, etc.
BioBrick BBa_T9002
is an HSL biosensor, which provides a non linear relationship between
HSL input and Scell output. More precisely, the
characteristic sigmoidal curve requires synthetic parameters for its
accurate identification. These are the minimum and maximum values, the
swtich point (i.e., the curve inflection point), and the upper and
lower boundaries of linearity. This biosensor revealed greatly
reliable, providing measurement repeatability and minimal experimental
noise. Referring to its activation formula, the calibration curve is
shown below.
In order to determine the threshold sensitivity of
T9002 biosensor, experiments were performed with several HSL inductions
minimally interspaced in the region of low detectability. Hypothesizing
that the inducer is 1:20 diluted (as for all of our tests), the minimum
detectable HSL concentration is 3 nM.
This biosensor was used to measure HSL concentration for parts producing or degrading this signalling molecule. For each of these experiments, a calibration curve of the biosensor was built, inducing it with known HSL concentrations, evaluating for each of them the Scell signal and finally estimating the Hill curve parameters. Once identified the parameters, the unknown concentration of a sample can be evaluated from its Scell (provided that it has a value included in the linear zone of the biosensor), as shown below: It is necessary, then, to multiply the measurement for the dilution factor used (in our experiments it was 20). |
|
•••
iGEM_Aberdeen_2014 |
Team Aberdeen iGEM 2014 Characterisation of BioBrick T9002This BioBrick is a GFP Expresser Controlled by 3OC6HSL Receiver Device. BioBrick BBa_T9002 is a composite part encoding a quorum sensing (QS) receiver driving expression of green fluorescent protein.
Aims and RationaleT9002 was previously reported to exhibit increased GFP expression in the presence of AHL (N-acyl homoserine lactone). We aimed to confirm GFP-expression responsiveness and dependence of AHL, and how the physical proximity of this QS receiver to any QS sender (producer of homoserine lactone) is facilitated by surface-binding affects quorum signalling. Materials and MethodsT9002 on a pSB1C3 backbone was rescued from Spring 2014 Distribution Plate 3-22J and transformed into XL1Blue E. coli competent cells, and selected on Chloramphenicol LB agar. Chloramphenicol LB liquid was innoculated with T9002 E. coli transformant and these were grown in conjunction with other E. coli transformed with the AHL ‘Sender’ plasmid Part:BBa_K1090000 (also on pSB1C3). BBa_K1090000 codes for AHL-synthesising enzymes, and also constitutively expresses RFP. Poly-L-lysine Cell AdhesionTo investigate QS signalling, and how it is affected by binding of sender and receiver bacteria to a solid surface, we immobilised our cells on a surface coated with poly-lysine:
ResultsK1090000 RFP expression RFP expression of K1090000 transformed E. coli was compared against untransformed XL1-Blue E. coli negative control and an RFP pSB1C3 plasmid positive control. Analysis of red fluorescence using a fluorimeter plate reader clearly shows that K1090000 constitutively expressed moderate amounts of RFP (Figure 1). T9002 with AHLT9002 E. coli was resuspended in conditioned growth medium containing AHL, derived from a filter sterilised K1090000 suspension. The T9002 transformants in this medium as expected displayed a slow, exponential increase in GFP, indicating the T9002 receiver cells were responding to the QS signalling caused by AHL in the medium. However, QS responses by the T9002 receivers were even more marked when T9002 transformants were co-cultured with K109000 QS sender transformants (Figure 2). Figure 1 - K1090000 E. coli constitutively expresses RFP. Red (Excitation 544nm/Emission 612nm) fluorescence of K1090000 E. coli (triangles), RFP positive control pSB1C3 (Circles), untransformed XL1-Blue E. coli (dashes).
Figure 2 – Production of GFP by T9002, induced either by filtered AHL-containing culture medium, or by actively growing K1090000 sender transformants. AHL derived from culture medium is sufficient for slow rate T9002 Receiver GFP production (-), although T9002 GFP production was more efficient when paired with actively-growing K1090000 senders (circles). Mean GFP fluorescence of E.coli free in 50µl suspensions incubated at 37C, 1mm double-orbital shaking for 30 seconds every 5 minutes for 355 minutes. Figure 3 – Greatest absolute fluorescence by T9002 transformants is observed in a Sender (S) 1:100 Receiver (R) cell number initial ratio. T9002 Receiver-produced GFP was compared with that produced when paired with an untransformed XL1-Blue E. coli (X) control or K1090000 Senders (circles), in triplicate. 50µl suspensions were incubated at 37C, 1mm double-orbital shaking for 30 seconds every 5 minutes for 355 minutes. Figure 4 – K1090000 Sender E. coli induction increased GFP expression in T9002 Receivers. Growth started in cell number ratios of aliquots totalling OD600 0.02 densities, suspended in liquid LB. Untransformed XL1-Blue:Receiver 1:100 ratio (Filled Diamonds), Sender:Receiver 1:100 ratio (Open circles). Values are blank corrected. T9002 and K1090000 bound to a Poly-Lysine surface. Overall, immobilised Sender/Receiver pairs exhibited a greater extended rate of GFP production response, and higher absolute response after 7 hours than sender-receiver pairs free in suspension; Figure 5. Figure 5: Surface-immobilisation of sender-receiver QS tranformants results in improved GFP expression. Poly-L-lysine wells (triangles) had greater rate of response compared to cells free in suspension (circles). This was performed in a Sender 1:10 Receiver cell number ratio.
Conclusions
|
|
••••
iGEM_Stockholm_2015 |
Method: For each experiment fresh solutions in was prepared in ddH2O in the following concentrations: 100 mM; 10 mM; 0.1 mM; 0.01 mM. We tested these different concentrations in 3 repetitions first in agar plates with the bacteria with BBa_T9002. Then we tested these different concentrations in the liquid LB cultures with 3 repetitions for each sample. Both plates and liquid cultures were incubated in 37°C for 24h. Results: No color change was observed for any of the experiments. It seems that our bacterial strain did not respond to any concentrations of OHHL in agar or liquid cultures. Conclusion: Based on our results, this part of our experiment did not work as intended. Please, visit our [http://2015.igem.org/Team:Stockholm iGEM Stockholm 2015 webpage] for more details or contact us. |