Difference between revisions of "Part:BBa K1724000"
(→Works Cited) |
|||
| (9 intermediate revisions by one other user not shown) | |||
| Line 35: | Line 35: | ||
== Contribution of FDR-HB_Peru 2022 == | == Contribution of FDR-HB_Peru 2022 == | ||
| − | In the parts registry of BBa_K1724000, we noticed that | + | In the parts registry of BBa_K1724000, we noticed that iGEM15_SCUT used 0.00003mM, 0.0001mM, 0.001mM, 0.01mM, 0.1mM as their cadmium ions concentration to test their cadmium biosensor with pcadA, a promoter that our team is also aiming to use in the future. Hence, we were curious to see if the E. coli is able to survive at higher concentrations than the ones tested in the registry. Additionally, “E. coli is intrinsically tolerant to high levels of cadmium up to 0.9–1.0 mM” (Qin et al.). Subsequently, we decided to test cadmium resistance of E. coli at final concentrations of 0mM, 0.3mM, 0.6mM, and 1.2mM using the Kirby Bauer method. |
From our results, we found that DH5 alpha E. coli was resistant to cadmium concentrations from 0.3 to 0.9mM, as no zones of inhibition were identified. We also found that the bacteria was tolerant to 1.2mM of CdSO4 as well, and small colonies were found. This suggests that E. coli could tolerate cadmium concentrations up to 1.2mM, which is 0.2mM more than what (Qin et al.).suggested. Hence, we were able to conclude that engineered DH5 alpha E. coli could be used at cadmium concentrations up to 1.2mM for cadmium detection and accumulation. | From our results, we found that DH5 alpha E. coli was resistant to cadmium concentrations from 0.3 to 0.9mM, as no zones of inhibition were identified. We also found that the bacteria was tolerant to 1.2mM of CdSO4 as well, and small colonies were found. This suggests that E. coli could tolerate cadmium concentrations up to 1.2mM, which is 0.2mM more than what (Qin et al.).suggested. Hence, we were able to conclude that engineered DH5 alpha E. coli could be used at cadmium concentrations up to 1.2mM for cadmium detection and accumulation. | ||
| + | |||
| + | |||
| + | [[File:cd resistance.png|200px|]] | ||
| + | |||
| + | (figure 1. One of the streaked plates for the experiment) | ||
=== Works Cited=== | === Works Cited=== | ||
| Line 47: | Line 52: | ||
[[Image:2015result-specific.png|500px|thumb|center|Figure 6. The specificity of the MerR/CadA promoter. As seen in the picture, the medium added cadmium in became red, in contrast that the medium which was added nothing or added other heavy metal maintained the same situation. ]] | [[Image:2015result-specific.png|500px|thumb|center|Figure 6. The specificity of the MerR/CadA promoter. As seen in the picture, the medium added cadmium in became red, in contrast that the medium which was added nothing or added other heavy metal maintained the same situation. ]] | ||
| + | |||
| + | ==Contribution== | ||
| + | *'''Group:''' iGEM24_HongKong-JSS | ||
| + | |||
| + | |||
| + | ===Cadmium biosensor=== | ||
| + | |||
| + | Our team's project aims to create biosensors for various heavy metals. Inspired by this part, we designed a biosensor (<partinfo>BBa_K5152006</partinfo>) using this promoter and the modified MerR. | ||
| + | |||
| + | We demonstrated that in our design, the biosensor responded to a 200 µM concentration of cadmium concentration by expressing a blue chromoprotein amilCP (<partinfo>BBa_K592009</partinfo>) as the reporter signal. This further validates the functionality of this promoter in response to cadmium and its potential use in different construct designs. | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <img src="https://static.igem.wiki/teams/5152/part-registry/17ab-merr-pcada-functional.webp" alt="100 uM Pb 12 hours" width="600"> | ||
| + | <figcaption><u>Fig. 7: Our biosensor exposed to 200 µM cadmium (II) chloride displayed an observable blue colour in the pellets and a less noticeable effect in the culture. </u> </figcaption> | ||
| + | </center> | ||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 11:39, 28 September 2024
Pcada
Introduction
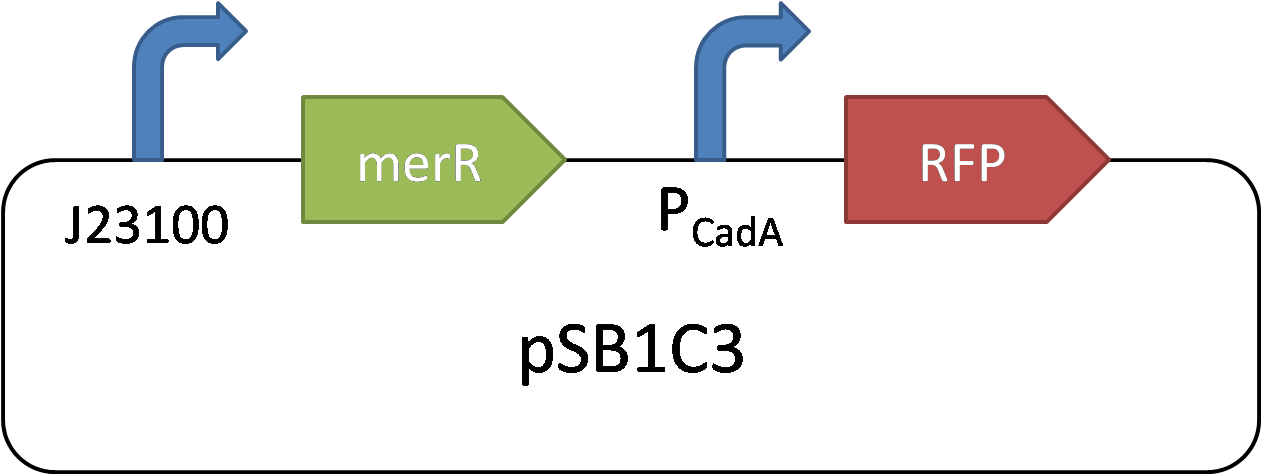
CadA promoter is a cadmium-sensitive promoter. Its repressor is MerR. In high concentration of cadmium, the Cd2+ can rapidly bind with MerR and remove its inhibition of CadA promoter. The reverse is the opposition.
Comparing with previous parts about cadmium sensors
1.Promoter CadA quantification
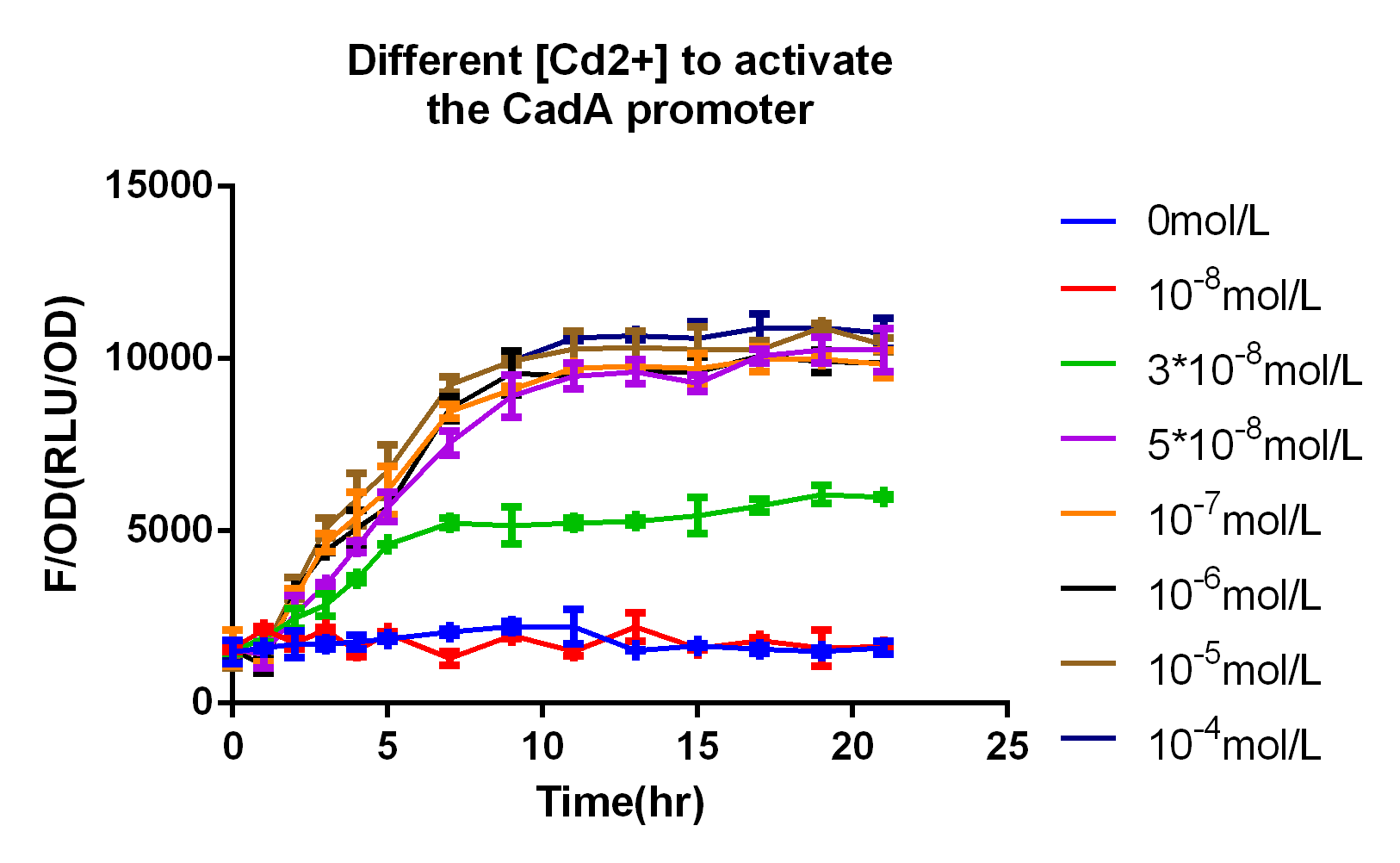
In 2015 SCUT team, we expressed our three proteins by the downstream of promoter CadA. Promoter CadA is Cd-activated promoter with the presence of MerR. When enough Cadmium existed, the gene downstream can be express. We construct MerR/CadA operon and place the RFP behind the CadA promoter. As the detection limitation mentioned below, the lowest concentration of the cadmium ion is between 10^-8 and 3*10^-8. So we set up the gradient concentration(0, 3*10^-8,10^-7,10^-6,10^-5,10^-4) of the cadmium ion in order to get the pattern of our promoter.

2.Detection limit of promoter CadA
In order to test the detection limit of the promoter CadA, we first set up gradient concentration of Cadmium(10^-8,10^-7,10^-6,10^-5,10^-4mol/L)to estimate the probably range. As the result mentioned above, the promoter CadA can be activated over the concentration of 3*10^-8. So we set up the gradient of Cd2+ (3*10^-9, 10^-8, 3*10^-8, 5*10^-8) and obtain the detection limit of promoter CadA.


Contribution of FDR-HB_Peru 2022
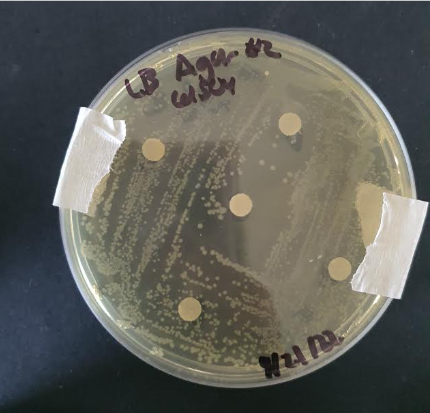
In the parts registry of BBa_K1724000, we noticed that iGEM15_SCUT used 0.00003mM, 0.0001mM, 0.001mM, 0.01mM, 0.1mM as their cadmium ions concentration to test their cadmium biosensor with pcadA, a promoter that our team is also aiming to use in the future. Hence, we were curious to see if the E. coli is able to survive at higher concentrations than the ones tested in the registry. Additionally, “E. coli is intrinsically tolerant to high levels of cadmium up to 0.9–1.0 mM” (Qin et al.). Subsequently, we decided to test cadmium resistance of E. coli at final concentrations of 0mM, 0.3mM, 0.6mM, and 1.2mM using the Kirby Bauer method.
From our results, we found that DH5 alpha E. coli was resistant to cadmium concentrations from 0.3 to 0.9mM, as no zones of inhibition were identified. We also found that the bacteria was tolerant to 1.2mM of CdSO4 as well, and small colonies were found. This suggests that E. coli could tolerate cadmium concentrations up to 1.2mM, which is 0.2mM more than what (Qin et al.).suggested. Hence, we were able to conclude that engineered DH5 alpha E. coli could be used at cadmium concentrations up to 1.2mM for cadmium detection and accumulation.
(figure 1. One of the streaked plates for the experiment)
Works Cited
Qin, Weitong, et al. Identification of Cadmium Resistance and Adsorption Gene from Escherichia Coli BL21 (DE3) - RSC Advances (RSC Publishing) DOI:10.1039/C7RA10656D. Royal Society of Chemistry, 6 Nov. 2017, pubs.rsc.org/en/content/articlehtml/2017/ra/c7ra10656d.
3. The specificity of the CadA promoter
Contribution
- Group: iGEM24_HongKong-JSS
Cadmium biosensor
Our team's project aims to create biosensors for various heavy metals. Inspired by this part, we designed a biosensor (BBa_K5152006) using this promoter and the modified MerR.
We demonstrated that in our design, the biosensor responded to a 200 µM concentration of cadmium concentration by expressing a blue chromoprotein amilCP (BBa_K592009) as the reporter signal. This further validates the functionality of this promoter in response to cadmium and its potential use in different construct designs.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]