Difference between revisions of "Part:BBa K302033"
(→Source and Principle) |
(→The verification of heat triggered cell lysis and suicide) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 104: | Line 104: | ||
[[File:XJTU-p5-2.png|500px]] | [[File:XJTU-p5-2.png|500px]] | ||
| + | |||
Figure 2:The principle of temperature-controlled suicide circuit | Figure 2:The principle of temperature-controlled suicide circuit | ||
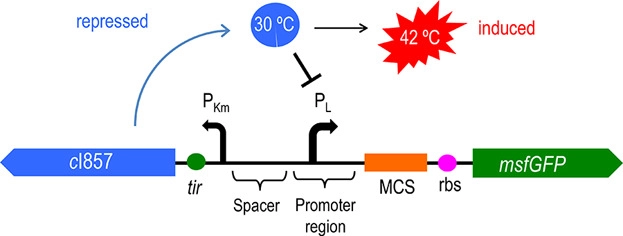
Figure 2 shows the principle of our temperature-controlled suicide circuit. When bacteria grows at a low temperature(30℃), the CI857 protein binds to the Pλ promoter, and downstream lysis gene are unable to be expressed, allowing the cell growth. While at 42℃, the CI protein will be degraded and lead to the expression of lysis gene and eventually cell death and release of the product. | Figure 2 shows the principle of our temperature-controlled suicide circuit. When bacteria grows at a low temperature(30℃), the CI857 protein binds to the Pλ promoter, and downstream lysis gene are unable to be expressed, allowing the cell growth. While at 42℃, the CI protein will be degraded and lead to the expression of lysis gene and eventually cell death and release of the product. | ||
| − | ===Codon | + | ===Codon optimization=== |
| − | The | + | The lysis gene is from Enterobacteria phage KleenX174. To better express the lysis gene in engineered bacteria, the codon optimization of the lysis gene was conducted according to the codon preference of Escherichia coli. Figure 3 shows the number of codons we optimized to make our codons more in line with Escherichia coli preference. The modified lysis gene is shown in BBa_K4182007. |
[[File:XJTU-p5-3.png|500px]] | [[File:XJTU-p5-3.png|500px]] | ||
| − | Figure 3: | + | Figure 3: Codon Optimization of lysis gene |
| − | ===Optimization of | + | ===Optimization of suicide circuit=== |
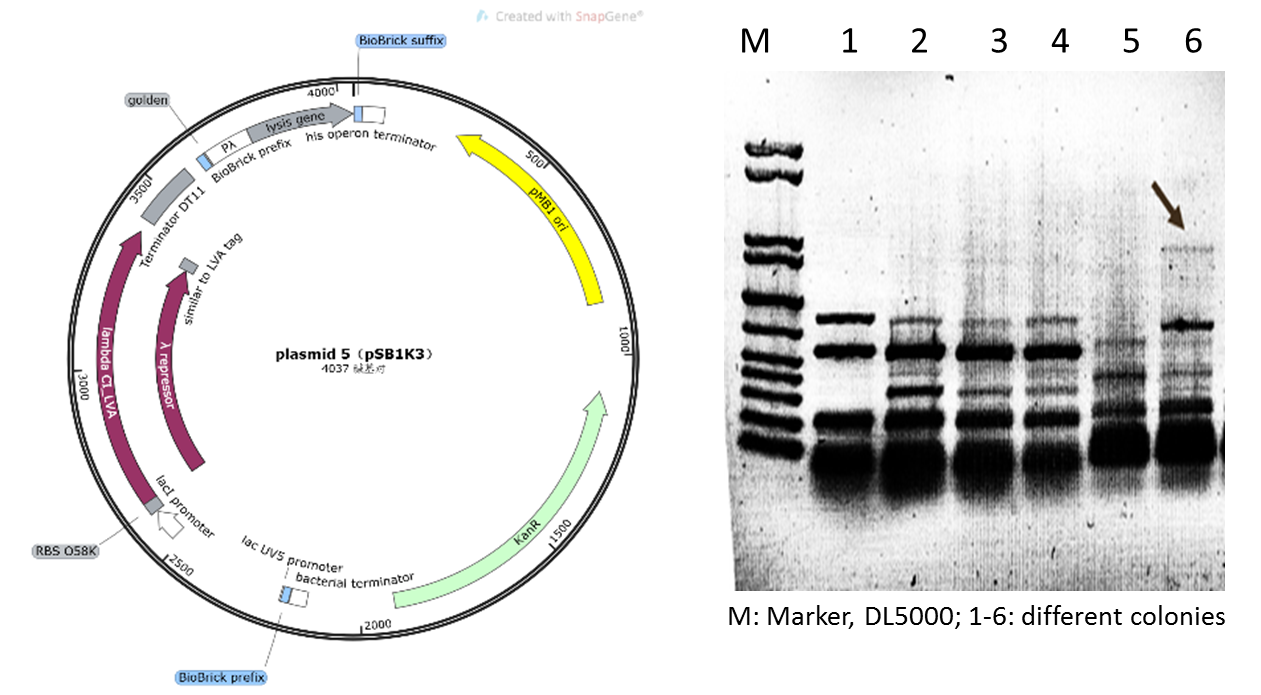
| − | Initially | + | Initially, we planned to construct our suicide circuit (Plasmid 5) using vector backbone pSB1K3 by Golden Gate assembly, and we can obtain several clones. However, it was found that after colony PCR verification, only weak target bands could be observed (Figure 4), and the plasmids extracted from the recombinant DH5α cells was at very low concentration, and sequencing could not be completed. |
[[File:XJTU-p5-10.png|500px]] | [[File:XJTU-p5-10.png|500px]] | ||
| Line 124: | Line 125: | ||
Figure 4: Plasmid 5 map based on pSB1K3 and its verification (he target band is approximately 1600bp) | Figure 4: Plasmid 5 map based on pSB1K3 and its verification (he target band is approximately 1600bp) | ||
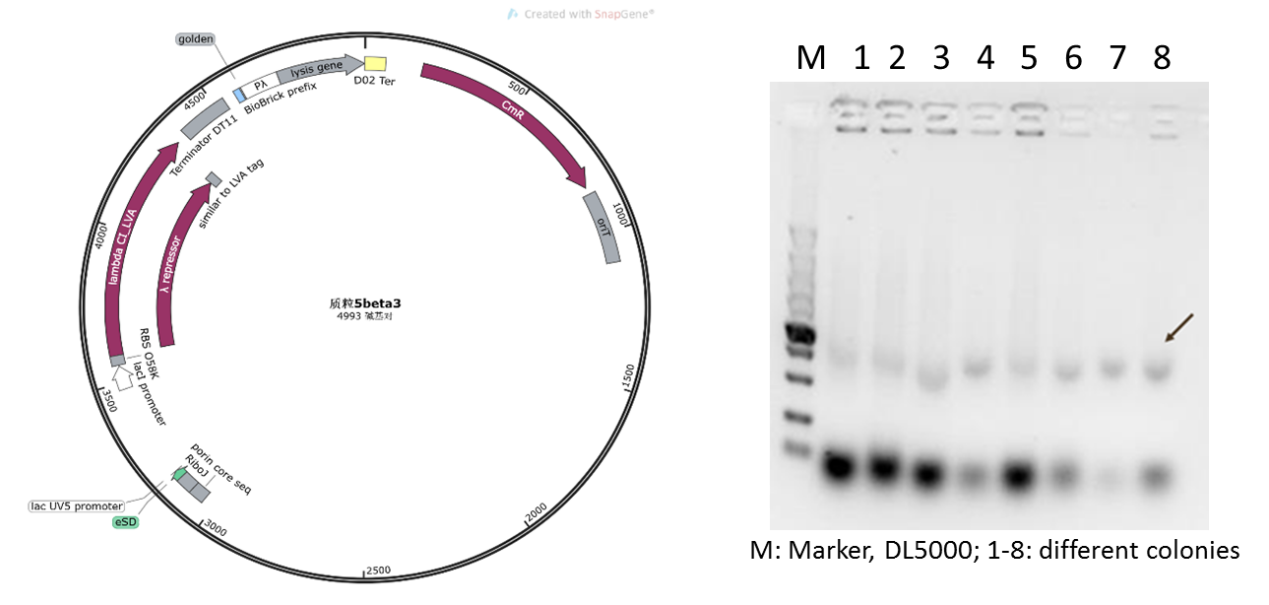
| − | + | We supposed that due to the low copy number of pSB1K3 vector, it is difficult to extract sufficient plasmid from the engineered bacteria for further validation. Therefore, we replaced the backbone to pSEVA341, a higher-copy-number vector and re-constructed the plasmid (Figure 5). As shown in Figure 5, the obvious target bands were observed, and the plasmid correctness was further confirmed by sequencing. | |
| − | As shown in Figure | + | |
[[File:XJTU-p5-11.png|500px]] | [[File:XJTU-p5-11.png|500px]] | ||
| − | Figure 5: new plasmid 5 with pSEVA341 backbone and its verification | + | Figure 5: The new plasmid 5 with pSEVA341 backbone and its verification |
===The verification of heat triggered cell lysis and suicide=== | ===The verification of heat triggered cell lysis and suicide=== | ||
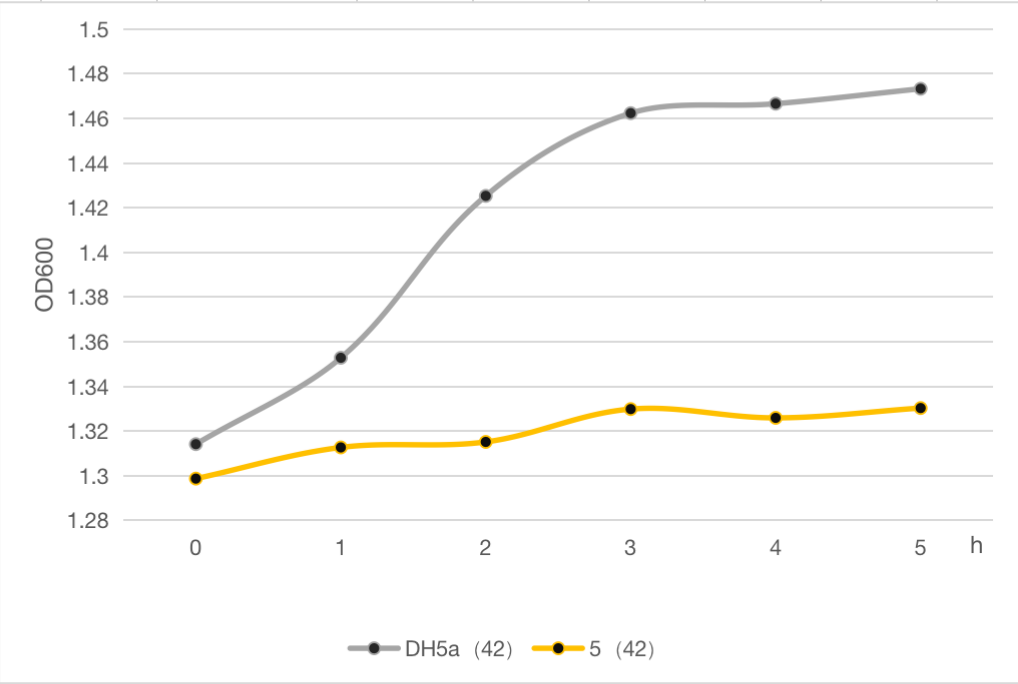
| − | The engineered cell harboring plasmid 5 and blank vector respectively, were culture at 30℃ overnight, and then the temperature was shift to 42℃ | + | The engineered cell harboring plasmid 5 and blank vector respectively, were culture at 30℃ overnight, and then the temperature was shift to 42℃. The OD600 of each group was detected every 1 h, and the growth curve of these strains were determined as follows. |
| + | The results clearly demonstrated the cell growth was significantly inhibited after heat at 42℃ compared to the strain without lysis gene. And about 9% of whole cell population was retained after heat. Compared to the commonly used suicide protein MazF (BBa_K302033), the lysis protein in our study is also efficient but shorter and easy to be manipulated, which can be used as an alternative and update for MazF. | ||
| − | |||
[[File:XJTU-p5-12.png|500px]] | [[File:XJTU-p5-12.png|500px]] | ||
Latest revision as of 18:26, 13 October 2022
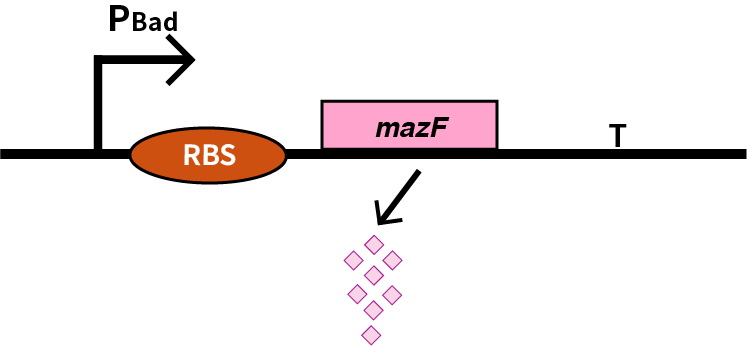
mazF
Encodes a stable non-specific ribonuclease in Bacillus Subtilis. It is used in conjungtion with mazE in Bacillus Subtilis to provide a toxin-antitoxin kill switch in various stressful conditions. When expression of both genes is turned off (as both are under contol of same promoter) mazE will be degraded faster than mazF. There is then no inhibiton of mazF, killing the cell.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterized by BNU-China 2019
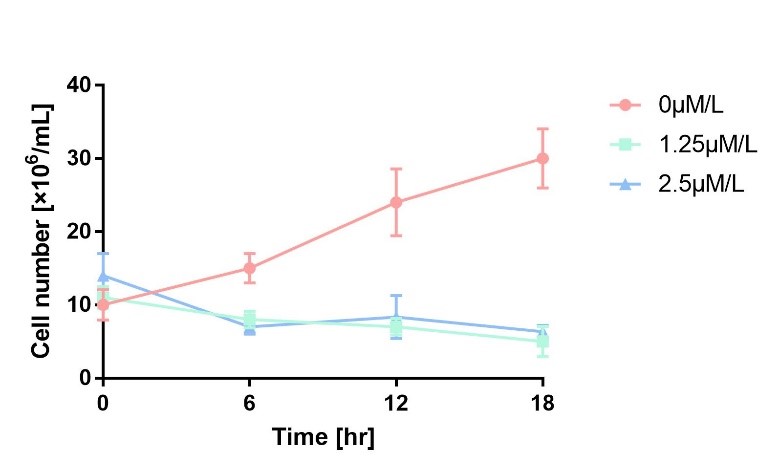
We characterize mazF (BBa_K302033) by an induced suicide system, in which the downstream mazF (BBa_K302033) gene encoding toxin MazF is put under the control of L-arabinose induced promoter pBAD (BBa_K206000). MazF is an endoribonuclease that cleaves RNAs at ACA sites and causes the death of microbe as a reporter [1]. As a result, we can characterize mazF in a cell density-dependent manner in Escherichia coli K-12.
In order to characterize the toxicity of protein encoded by mazF, we take engineered microbe without induction as control group. As is shown in Fig.1, the cell number of experimental groups show a significant decrease upon induction, which indicates the protein encoded by mazF is lethal to E. coli.
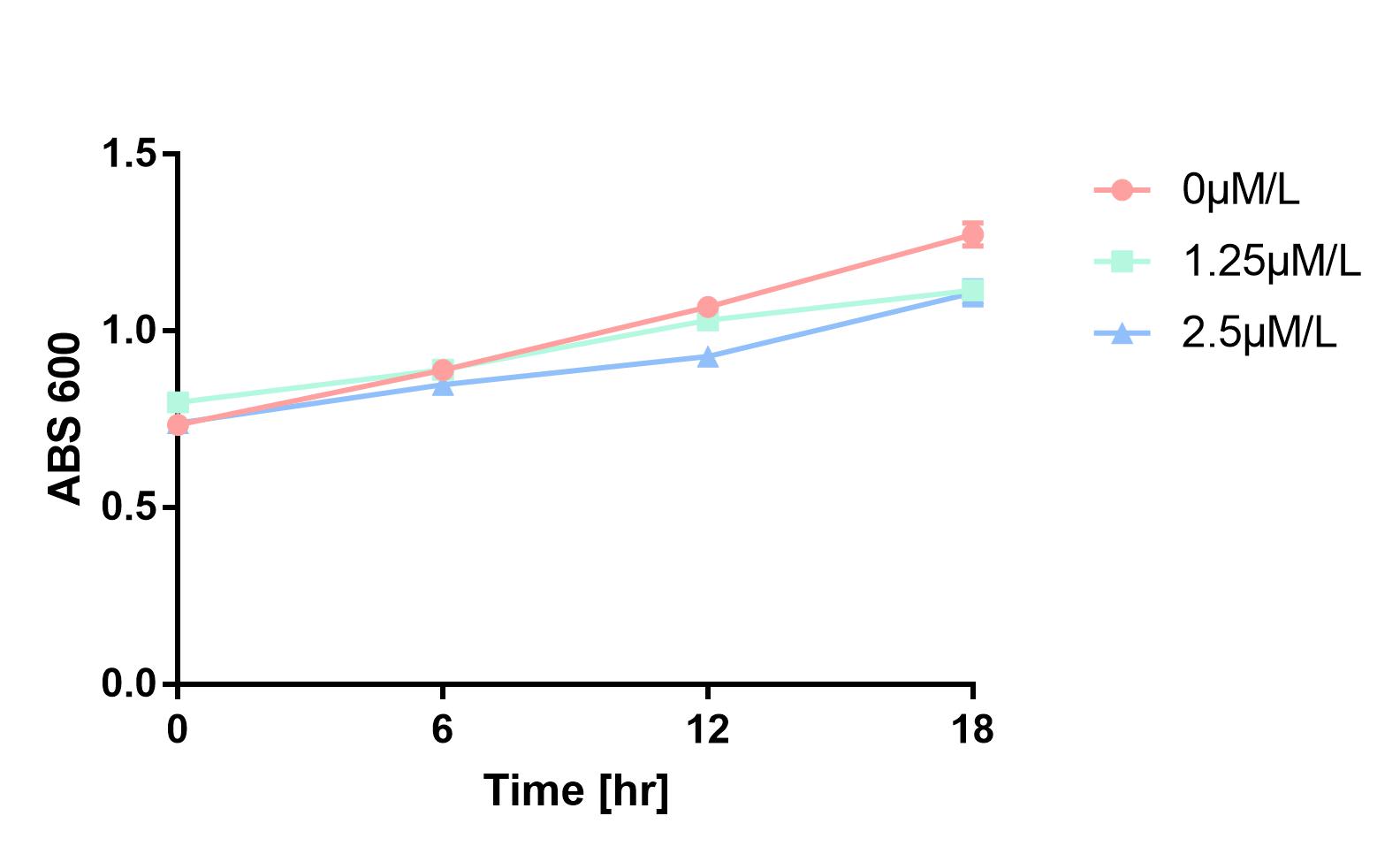
Furthermore, in order to prove that MazF kills the cell without lysing it, we measure OD600 of each sample to give an overall number of intact bacteria, dead and alive. As is shown in Fig.2, there is little difference between control and experimental groups, although it is validated numbers of alive cells differ. Hence, we reach a conclusion that protein encoded by mazF mediates suicide of cells without causing lysis of bacterial cells.
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells.
2. The engineered bacteria are cultured in 200mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm;
3. Equally divide the culture into 90 centrifuge tubes, which is 1mL respectively. Centrifuge them at 4000rpm for 5 minutes. Discard the liquid.
4. Resuspend 30 tubes of collected bacteria with LB-ampicillin (50 ng/µl) containing 1.25μM/L and 2.5μM/L L-arabinose respectively as experimental groups. Resuspend 30 tubes of bacteria with pure LB-ampicillin (50 ng/µl) medium.
5. Collect 3 tubes of all groups every 6 hours, dilute all of the samples to 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately. At the same time, refresh the medium to maintain the concentration of L-arabinose.
6. Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃
7. Three repicas are tested in each group.
Reference
[1] Diana Széliová, Ján Krahulec, Martin Šafránek, et al. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains[J]. Journal of Biotechnology, 2016, 236:1-9.
Characterized by XJTU-China 2020
Characterization
This year, our team designed a suicide switch mediated by arabinose operon for biosafety. We use MazF to kill the bacteria. We have supplemented the relevant information of this element in the iGEM Parts and added test and characterization to it.
Our MazF comes from the genome of Bacillus Subtilis. By regulating the expression of MazF, we inhibit the life of bacteria when needed to prevent the possible strains and genetic leakage. MazF is a kind of mRNA interferas. Specifically, it specifically cleaves five-base U^ACAU sequence (^indicating the cleavage site) on RNA[1]. MazF achieves its catalytic function by forming a special MazF-RNA complex. One molecule of 9-mer RNA is bound to one MazF dimer, and two subunits of MazF form the dimer related by local 2-fold symmetry.The RNA in an extended alignment is bound along the RNA binding interface between subunits of the MazF dimer, covering part of this dimeric interface, and MazF interacts extensively with the pentad target sequence present in the RNA chain by interactions with bases[2]. Therefore, the backbone phosphate moieties project outward and away from the protein surface. In this way, scissile phosphate will be attacked by other side chain groups, which causes breaking of P-O bond.
What’s more, it is an important part of the Bacillus Subtilis toxin-antitoxin system. Which is essential for the programmed cell death of this developmental bacterium. In normally growing cells, MazF forms a stable complex with its cognate antitoxin, MazE, however, under stress conditions, unstable MazE is preferentially degraded to release free MazF in the cells, which then cleaves cellular mRNAs to inhibit protein synthesis, leading to growth arrest. The protein-protein interaction interface between MazE and MazF (2,843 Å2) is larger than the MazF-RNA interaction interface (2,153 Å2), suggesting that MazF is likely to have a higher affinity for MazE over its RNA substrate. In the B.S MazE-MazF complex structure, MazE binds to MazF along the dimer interface, and it even occupies part of the putative modeled RNA binding site on the second subunit of MazF[3]. Thus, in the presence of MazE, MazF cannot bind to or cleave substrate RNA.
Biochemical properties of MazF
divalent cation[4].
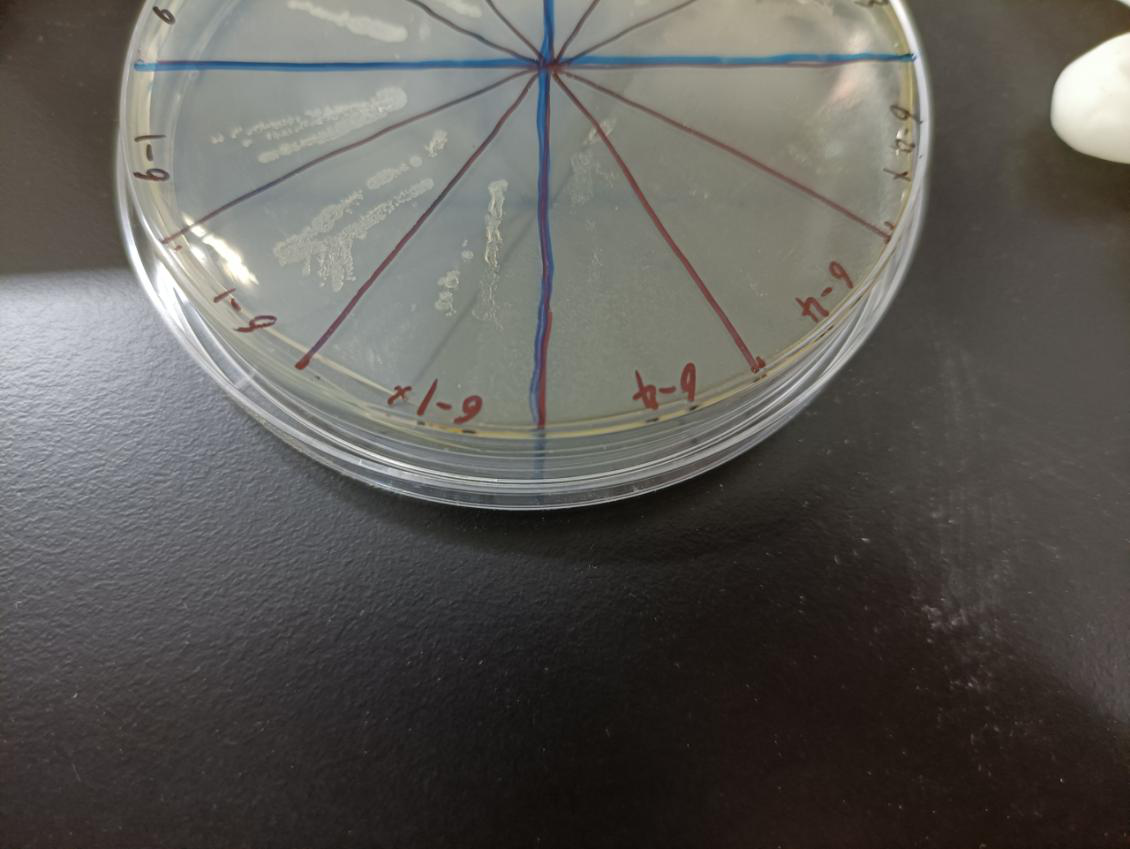
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 groups were the control group without arabinose induction, and three of 6-4 groups were the experimental group with 1.5 M/L arabinose induction. Where, the dilution factor of the group with * is 10^4, and the dilution factor of the group without * is 10^6.
Result: The bacteria without arabinose induction grow normally, while with arabinose induction, the bacteria suicide.
Reference
[1]Park, Jung-Ho, Yamaguchi, Yoshihiro and Inouye, Masayori(2011), Bacillus subtilis MazF‐bs (EndoA) is a UACAU‐specific mRNA interferase, FEBS Letters, 585, doi: 10.1016/j.febslet.2011.07.008 [2]Dhirendra K. Simanshu, Yoshihiro Yamaguchi, Jung-Ho Park, et al. Structural Basis of mRNA Recognition and Cleavage by Toxin MazF and Its Regulation by Antitoxin MazE in Bacillus subtilis. 2013, 52(3):447-458 [3]Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 2005; 56:845–857. [PubMed: 15853875] [4] Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol. 2005 Jun;56(5):1139-48. doi: 10.1111/j.1365-2958.2005.04606.x. PMID: 15882409.
Contribution & Improvement from XJTU-China 2022
Profile
Name
Lysis-phi X174
Base Pairs
273
Design Notes
The gene was optimized by E. coli codon
Source
Shigella flexneri 2a str. 301 (strain: 301, serotype: 2a)
Usage&Biology
Source and Principle
Biosafety is an important consideration when designing engineered bacteria, and MazF has been commonly used in previously work. Here we develop a novel lysis gene (BBa_K4182007) for the design of controlled suicide circuits for engineering efficiency and biosafety reasons, also can be considered as an alternative of suicide protein MazF.
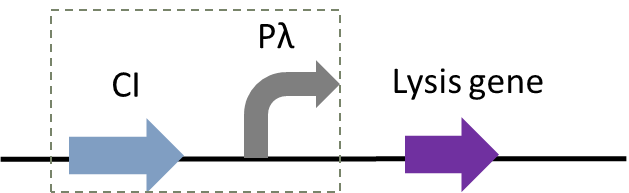
Figure 1: The temperature-controlled suicide circuit with a novel lysis gene
Figure 2:The principle of temperature-controlled suicide circuit
Figure 2 shows the principle of our temperature-controlled suicide circuit. When bacteria grows at a low temperature(30℃), the CI857 protein binds to the Pλ promoter, and downstream lysis gene are unable to be expressed, allowing the cell growth. While at 42℃, the CI protein will be degraded and lead to the expression of lysis gene and eventually cell death and release of the product.
Codon optimization
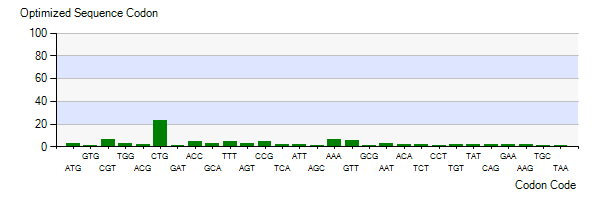
The lysis gene is from Enterobacteria phage KleenX174. To better express the lysis gene in engineered bacteria, the codon optimization of the lysis gene was conducted according to the codon preference of Escherichia coli. Figure 3 shows the number of codons we optimized to make our codons more in line with Escherichia coli preference. The modified lysis gene is shown in BBa_K4182007.
Figure 3: Codon Optimization of lysis gene
Optimization of suicide circuit
Initially, we planned to construct our suicide circuit (Plasmid 5) using vector backbone pSB1K3 by Golden Gate assembly, and we can obtain several clones. However, it was found that after colony PCR verification, only weak target bands could be observed (Figure 4), and the plasmids extracted from the recombinant DH5α cells was at very low concentration, and sequencing could not be completed.
Figure 4: Plasmid 5 map based on pSB1K3 and its verification (he target band is approximately 1600bp)
We supposed that due to the low copy number of pSB1K3 vector, it is difficult to extract sufficient plasmid from the engineered bacteria for further validation. Therefore, we replaced the backbone to pSEVA341, a higher-copy-number vector and re-constructed the plasmid (Figure 5). As shown in Figure 5, the obvious target bands were observed, and the plasmid correctness was further confirmed by sequencing.
Figure 5: The new plasmid 5 with pSEVA341 backbone and its verification
The verification of heat triggered cell lysis and suicide
The engineered cell harboring plasmid 5 and blank vector respectively, were culture at 30℃ overnight, and then the temperature was shift to 42℃. The OD600 of each group was detected every 1 h, and the growth curve of these strains were determined as follows. The results clearly demonstrated the cell growth was significantly inhibited after heat at 42℃ compared to the strain without lysis gene. And about 9% of whole cell population was retained after heat. Compared to the commonly used suicide protein MazF (BBa_K302033), the lysis protein in our study is also efficient but shorter and easy to be manipulated, which can be used as an alternative and update for MazF.
Figure 6: The cell growth of strains with suicide plasmid 5 and blank vector
References
1. Din, M.O., et al., Synchronized cycles of bacterial lysis for in vivo delivery. Nature, 2016. 536(7614): p. 81-85.
2. Saeidi, N., et al., Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol, 2011. 7: p. 521.
3. Restrepo-Pineda, S., et al., Thermoinducible expression system for producing recombinant proteins in Escherichia coli: advances and insights. FEMS Microbiol Rev, 2021. 45(6).
4. Aparicio, T., V. de Lorenzo, and E. Martínez-García, Improved Thermotolerance of Genome-Reduced Pseudomonas putida EM42 Enables Effective Functioning of the PL/cI857 System. Biotechnology Journal, 2019. 14(1): p. 1800483.
Characterized by BNUZH-China 2022
In this module, we replaced GOI with MazF. We hoped that when illuminated with the 660 nm light, MazF can be transcription and translation. Then MazF works, killing the engineered cells. Figure 1 illustrates the detailed design of the whole device.
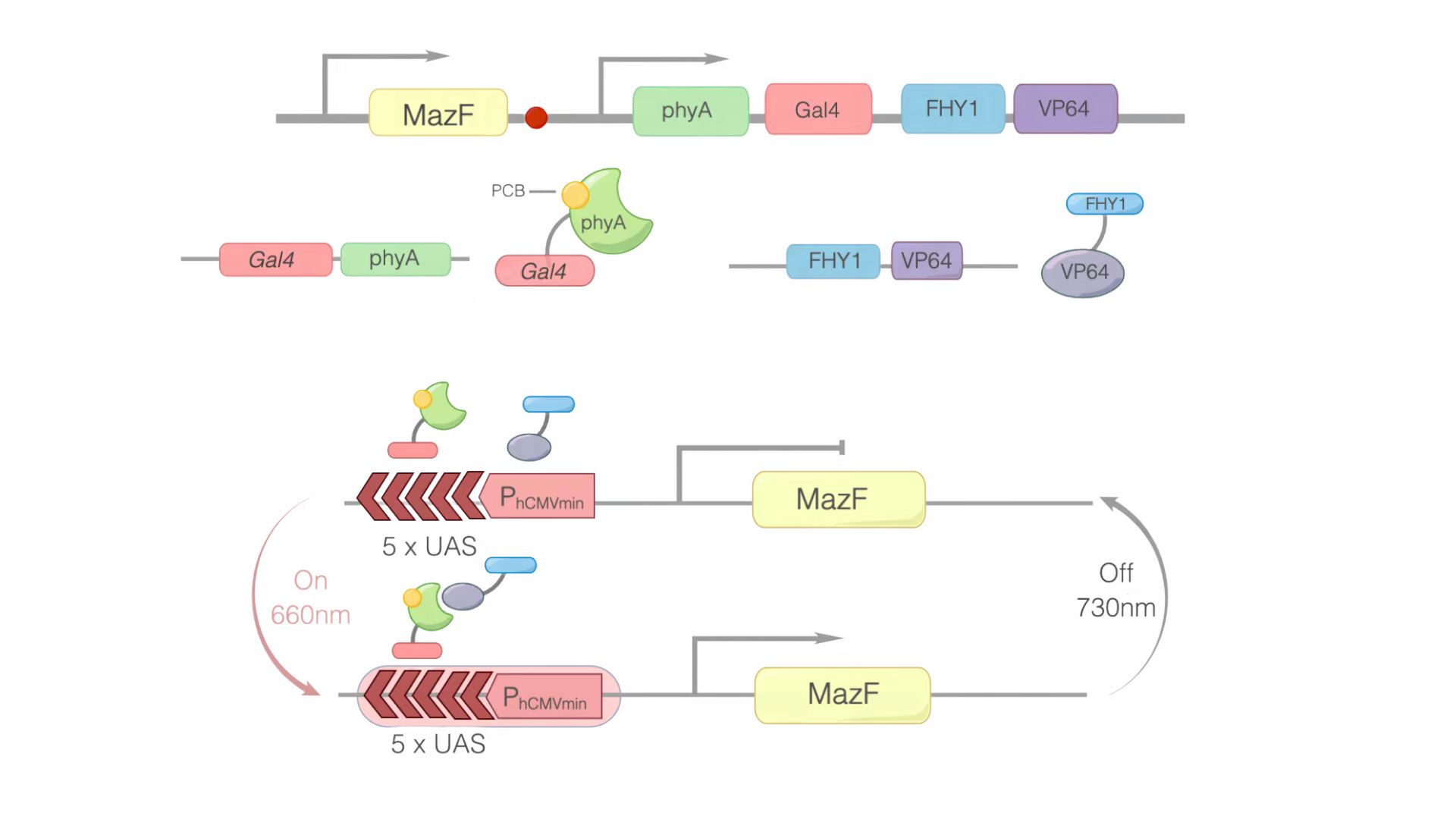
Figure 1: Construct design and the principle of the REDMAP-MazF. 5×UAS-P(hCMVmin) was fused to MazF. Gal4 was linked via a linker to PhyA. At the same time, PCB was combined with PhyA. PhyA turned to ΔPhyA. VP64 was linked with FHY1. When 660 nm light illuminating, ΔPhyA combines with VP64. Then, Gal4-ΔPhyA is transported into the nucleus by VP64-FHY1. After that, Gal4 will combine with 5×UAS-P(hCMVmin), activating the transcription of the downstream target gene: MazF. Then MazF works, killing the engineered cells.
Usage and Biology
We planned to use it to activate the transcription of MazF (BBa_K302033) to cause the engineered cells to commit suicide.
Experimental approach
For testing this device, we used HEK293T cells, which were seeded in 25cm2 flask. The PLVX vector was transiently transfected into HEK293T cells by Lipo8000™ transfection system and incubated for 43 h to ensure normal cell status. Then, phycocyanobilin (PCB) with a final concentration of 5 μM was added to the system, and the system was illuminated with red light (660 nm) for 3 h. After illumination, the cells were observed and the cell lysates were analyzed by SDS-PAGE and Western Blot.
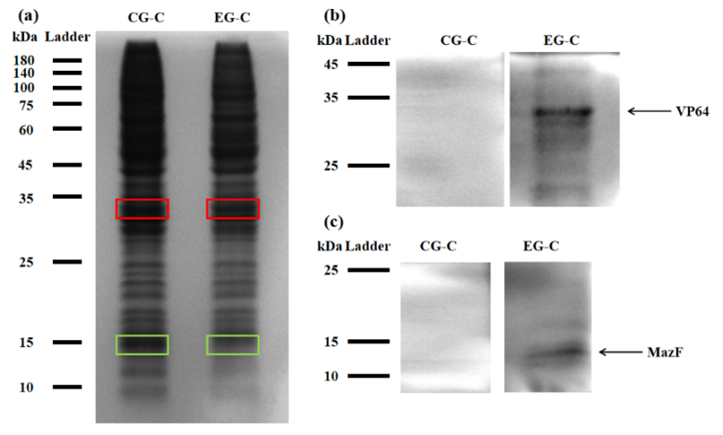
Figure 2: (a) REDMAP SDS-PAGE results. The VP64 strip is in the red box, and the MazF strip is in the green box. CG-C was the cell lysate of the control group, and EG-C was the cell lysate of the experimental group. (b) VP64 Western Blot diagram. CG-C was the cell lysate of the control group, EG-C was the cell lysate of the experimental group, and the theoretical size of VP64 was 30 kDa. (c) MazF Western Blot. CG-C was the cell lysate of the control group, EG-C was the cell lysate of the experimental group, and the theoretical MazF size was 13 kDa.
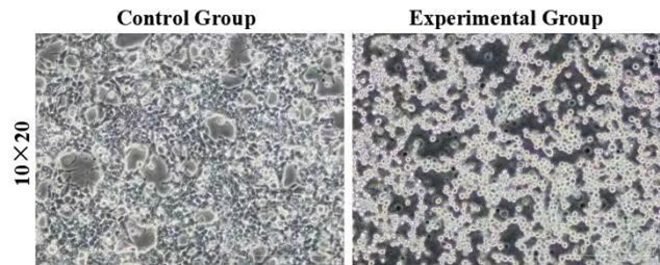
Figure 3: Morphology of control and experimental cells after red light verification. The control group was HEK293T cells without transient transfection, and the experimental group was HEK293T cells with transient transfection. 10×20 refer to the multiple of the eyepiece × the objective.
References
[1] Park, J. H., Yamaguchi, Y. & Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett 585, 2526-2532 (2011). https://doi.org:10.1016/j.febslet.2011.07.008
[2] Zhou, Y., Kong, D., Wang, X. et al. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat Biotechnol 40, 262–272 (2022). https://doi.org/10.1038/s41587-021-01036-w
[3] Simanshu, D. K., Yamaguchi, Y., Park, J. H., Inouye, M. & Patel, D. J. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol Cell 52, 447-458 (2013). https://doi.org:10.1016/j.molcel.2013.09.006
[4] Stein, T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56, 845-857 (2005). https://doi.org:10.1111/j.1365-2958.2005.04587.x
[5] Pellegrini, O., Mathy, N., Gogos, A., Shapiro, L. & Condon, C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol 56, 1139-1148 (2005). https://doi.org:10.1111/j.1365-2958.2005.04606.x
Improvement by 2022 Canton_HS
Autism Spectrum Disorder (ASD) is a neuro-developmental disorder characterized by social communication disorders. Our research aims at early diagnosis and accurately diagnosing and treatment. In order to carry forward the spirit of iGEM, we specially searched the iGEM Biological Parts library for related projects and picked BBa_K302033, MazF. This is a biological part submitted by iGEM10_Newcastle in 2010, they developed an ASD diagnosis and treatment method, but didn’t totally achieve ASD treatment through the brain-gut-axis. So that it is really important to provide another candidate protein for ASD treatment. In this project, our team carried out a related protein SLC7A4 for this part in the laboratory. The SLC7A5 transports the so-called branched-chain amino acids (BCAA) into the brain. Mutation in this gene reduces branched-chain amino acid levels in the brain and interferes with neural cell protein synthesis which reduced social interactions and other changes. Improving the SLC7A5 levels by building engineered probiotics may achieve the treatment of autism. In order to verify if the new part SLC7A5 we developed worked well, we transformed the recombinant plasmid into E. coli BL21(DE3) to verify the protein expression level by SDS-PAGE. This protein may could be used in future research.
Profile
Name: SLC7A5
Base Pairs: 1524 bp
Origin: Homo sapiens, genome
Properties: amino acid transporter
Usage and biology
Solute carrier transporter 7a5 (SLC7A5), also known as LAT1, belongs to the APC (amino acid, polyamine, choline) superfamily. The heterodimer SLC7A5 and glycoprotein CD98 linked through a conserved disulfide is responsible for the uptake of essential amino acids in crucial body districts such as the placenta and blood-brain barrier. The SLC7A5 transports the so-called branched-chain amino acids (BCAA) into the brain. Mutation in this gene reduces branched-chain amino acid levels in the brain and interferes with neural cell protein synthesis which shows reduced social interactions and other changes. There are many different genetic mutations causing autism, and this heterogeneity makes it difficult to develop effective treatments.
Construct design
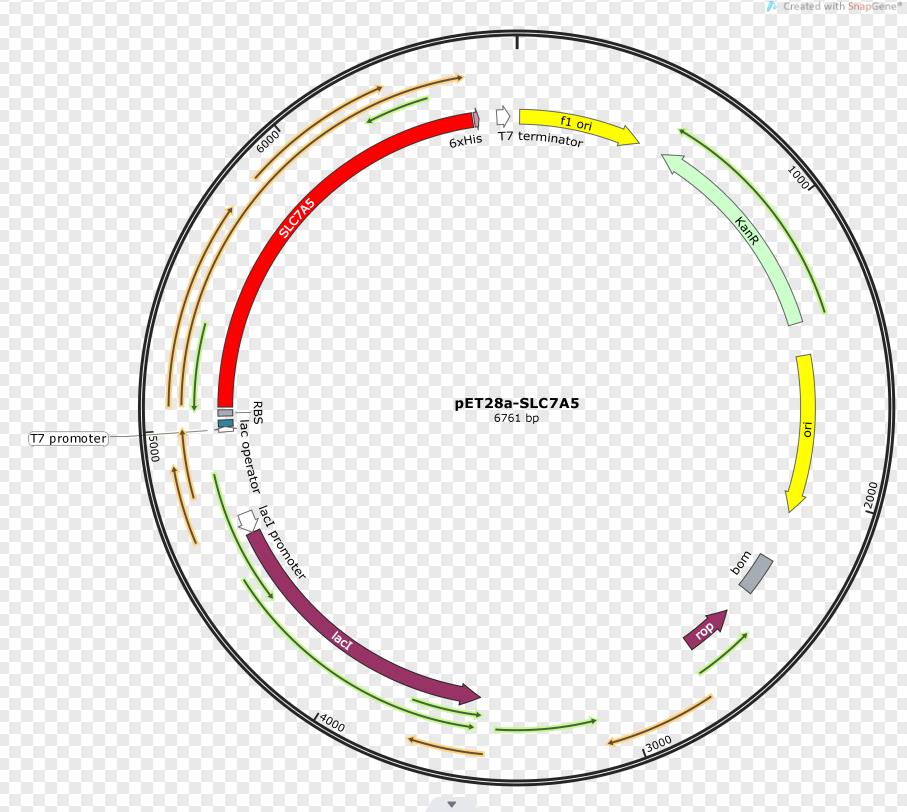
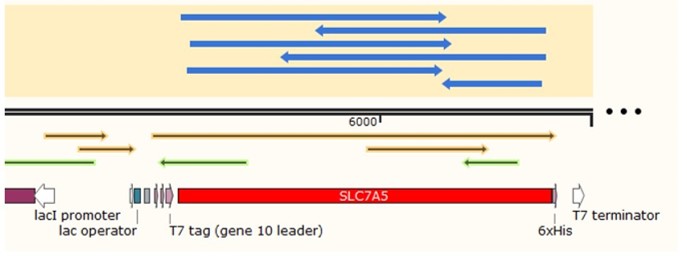
SLC7A5 transporter is expressed under the promoter T7. The gene fragment was inserted into pET28a.
Experimental approach
1.1 Construction of biosensor expression plasmids
In order to build our plasmids, we let the synthetic company synthesize the SLC7A5 DNA sequence and inserted it into the pUC57 vector. We amplified the SLC7A5 from the plasmid by PCR (Figure 2) and inserted it into the EcoRI and XhoI sites of the pET28a vector. We transformed it into E. coli Top10 competent cells.
1.2 Colony PCR of pET28a-SLC7A5
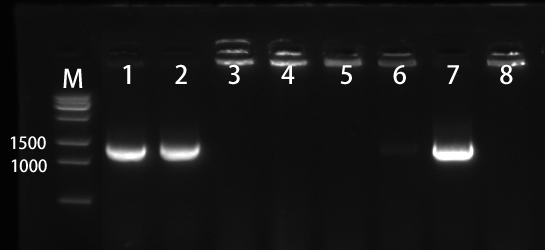
We selected the 8 colonies to perform by colony-PCR (Figure 3). The electrophoresis result showed that No. 1, 2, and 7 has correct bands.
1.2 Sanger sequencing of recombinant plasmid
We inoculated the correct colonies in LB (Kan+) liquid medium and extracted the plasmids and sent them to the company for Sanger sequencing. The returned sequencing data showed that there were no mutations in the gene region which means we successfully constructed the recombinant plasmid.
Proof of function
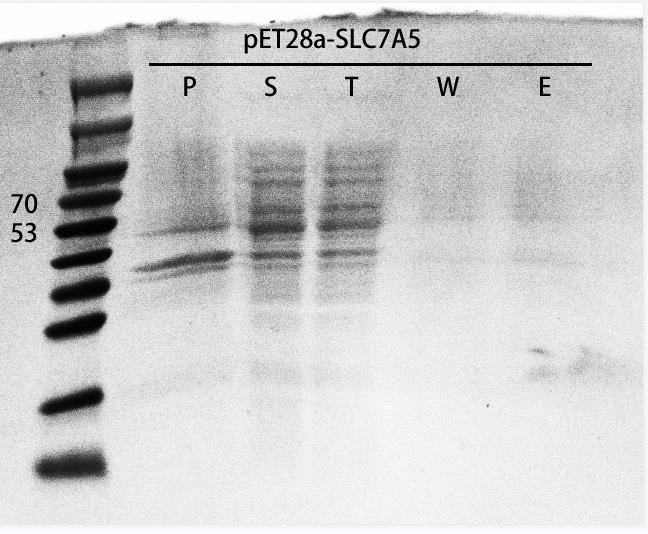
SLC7A5 purification
In order to verify if SLC7A5 expression in the E. coli host strain, we transformed the recombinant plasmids into E. coli BL21(DE3), inoculated the recombinants and added IPTG to induce protein expression when the OD600 reached 0.6. After overnight induce and culture, we collected the cells and ultrasonic fragmentation of cells to release the intracellular proteins. Next, we verified the expression level of the target protein by SDS-PAGE (Figure 5). As a result, the recombinants successfully expressed our target protein.