Difference between revisions of "Part:BBa K4274009"
| (18 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
==Characterization== | ==Characterization== | ||
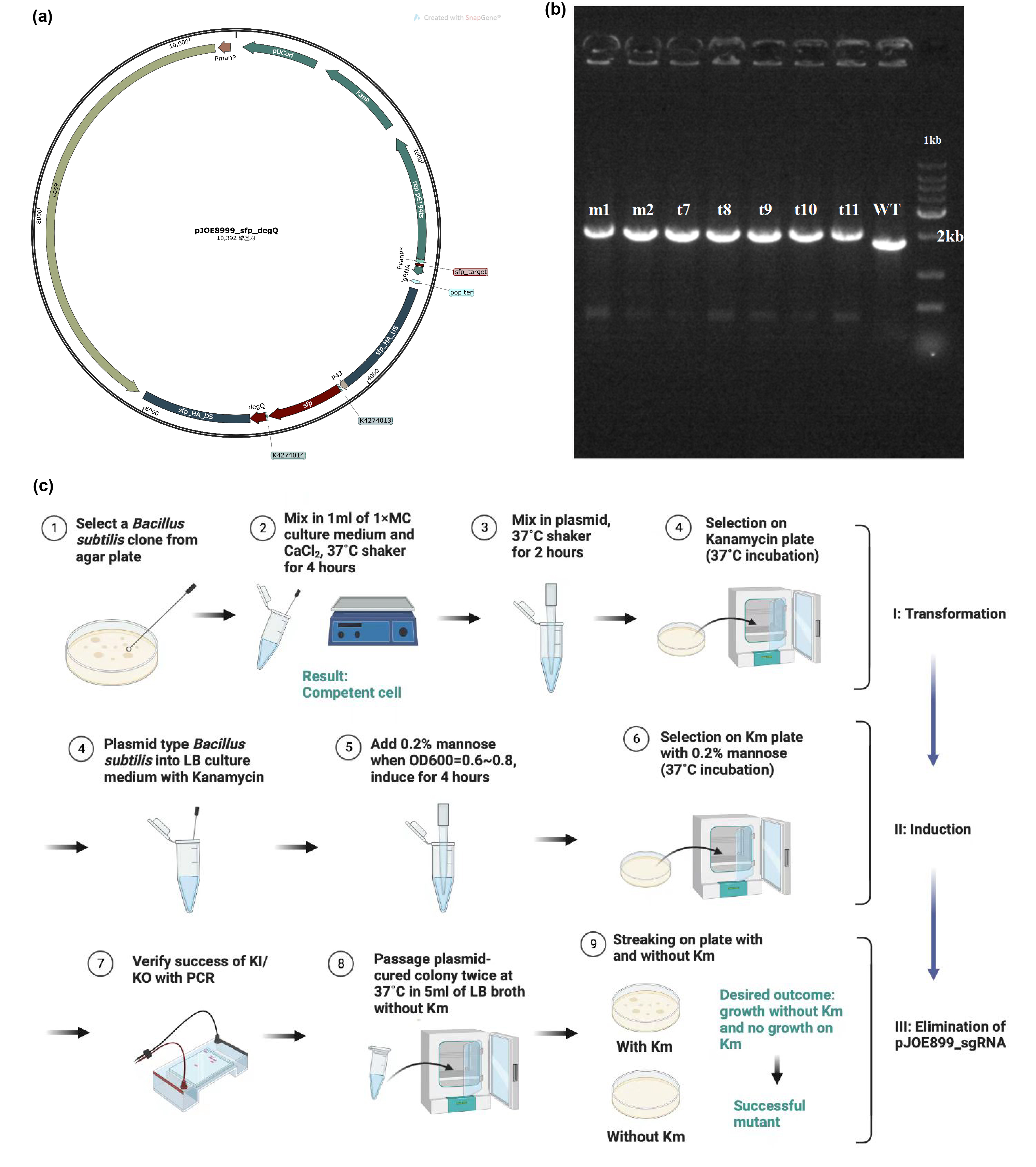
| + | The main composite of our product, fengycins, is produced by engineering <i>Bacillus subtilis</i> 168. As it is shown in the demonstration in description, sfp and degQ genes are critical to the biosynthesis of fengycins. Besides, <i>Bacillus subtilis</i> 168 possesses a natural but invalid sfp gene, and the functional degQ gene is silenced with the regulation of a weak promoter.Thus, with the help of pJOE8999_sfp_degQ plasmid (Figure 1a), we knocked in both sfp gene and degQ gene in the region of the natural sfp gene of <i>Bacillus subtilis</i> 168 after being knocking out. Under the guidance of protocol, we successfully transformed and induced the plasmid to function as a gene editor (Figure 1c). After induction, 7 strains were selected for PCR and electrophoresis verification. Since successful knock-in would would result in a 252 bps increase in the genome of <i>Bacillus subtilis</i> 168, the electrophoresis result conveys preliminarily that all of these strains has achieved our target of constructing the fengycins producers (Figure 1b). | ||
[[Image:Parts-keystone-fengycin1.jpeg|thumbnail|750px|center|'''Figure 1:''' | [[Image:Parts-keystone-fengycin1.jpeg|thumbnail|750px|center|'''Figure 1:''' | ||
Edting the genome of <i>Bacillus subtilis</i> 168 to enable it to produce fengycins. (a) Plasmid design for knocking out the invalid sfp gene of <i>Bacillus subtilis</i> 168 and knocking in the sfp gene from B. amyloliquefaciens FZB42 and degQ gene from <i>Bacillus subtilis</i> 168. (b) Electrophoresis results show that gene editing is successful. (c) Protocol about transformation, induction and elimination of pJOE8999 plasmid in <i>Bacillus subtilis</i> 168. ]] | Edting the genome of <i>Bacillus subtilis</i> 168 to enable it to produce fengycins. (a) Plasmid design for knocking out the invalid sfp gene of <i>Bacillus subtilis</i> 168 and knocking in the sfp gene from B. amyloliquefaciens FZB42 and degQ gene from <i>Bacillus subtilis</i> 168. (b) Electrophoresis results show that gene editing is successful. (c) Protocol about transformation, induction and elimination of pJOE8999 plasmid in <i>Bacillus subtilis</i> 168. ]] | ||
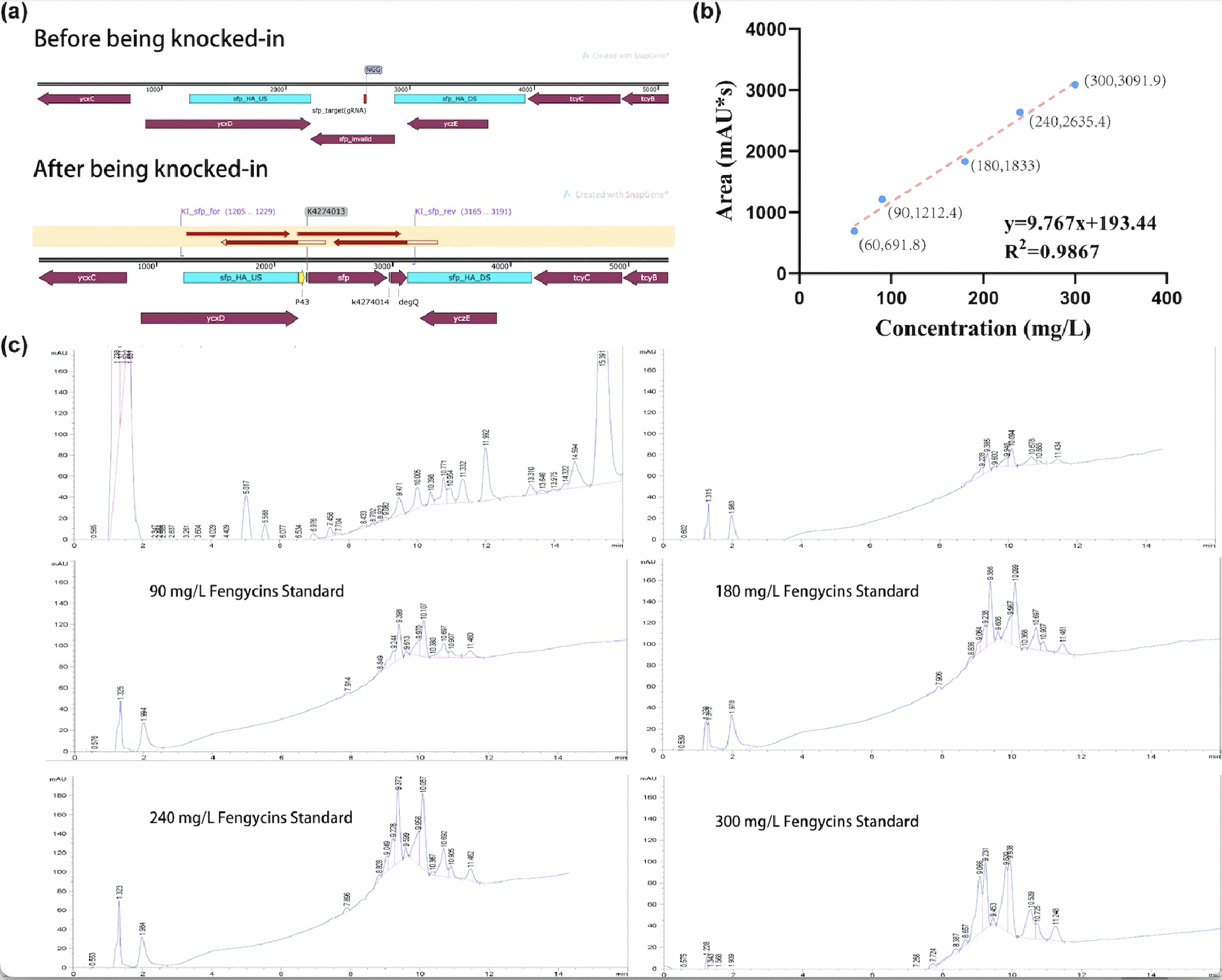
| − | + | After sequencing further verified that the successful construction of the fengycins producing strain, we used the strain for fermentation (Figure 2a). For the quantification of fengycins production, the methanol - extract obtained from the fermentation broth was analyzed by high performance liquid chromatography (HPLC). The peak of fengycins appeared at the retention time of 8-11.5 min under methanol mobile phase (Figure 2c). The injection sample was a solution of lipopeptide extract from 30 mL of fermentation broth dissolved in 7 mL methanol. Besides, various concentrations of fengycins standard is also injected for the construction of the standard curve of peak area obtained by HPLC (y) and its corresponding fengycins’ concentration (x). Based on the standard curve, we got the regression curve and its corresponding formula y=9.767x+193.44, and R2>0.98 displays that the regression equation fits the observed values (Figure 2b). Substituting y=1242.3 obtained from the sample into the regression equation, the concentration of fengycin in methanol solution can be calculated as 107.4 mg/L, that is, 25.06 mg/L of fengycins can be achieved by shaking-flask fermentation. | |
| − | [[Image:Parts-keystone- | + | [[Image:Parts-keystone-fengycin2-1.jpeg|750px|thumbnail|center|'''Figure 2:''' |
Quantification analysis of fengycins production. (a) The sequencing result of engineering <i>Bacillus subtilis</i> 168 strain for production. (b) The standard curve of peak area obtained by HPLC (y) and its corresponding fengycins' concentration (x). (c) HPLC results of sample and various concentrations of fengycins standard. ]] | Quantification analysis of fengycins production. (a) The sequencing result of engineering <i>Bacillus subtilis</i> 168 strain for production. (b) The standard curve of peak area obtained by HPLC (y) and its corresponding fengycins' concentration (x). (c) HPLC results of sample and various concentrations of fengycins standard. ]] | ||
| + | |||
==Source== | ==Source== | ||
<i>B. amyloliquefaciens</i> FZB42 | <i>B. amyloliquefaciens</i> FZB42 | ||
Latest revision as of 12:46, 12 October 2022
sfp
sfp gene is a biobrick part from B. amyloliquefaciens FZB42 encoding 4'-phosphopantetheinyl transferase which functions as a primer of nonribosomal peptide synthesis via phosphopantetheinylation of thiotemplates. However, compared to the sfp gene in Bacillus subtilis 168, they only had an amino acid homology of 70%. And the mutation existed in Bacillus subtilis 168 resulted in its unable to produce any lipopeptide biosurfactant. Hence, to allow B. subtilis to produce fengycins, it is essential to first knock-out the mutant gene and then knock-in the correct sfp (Part: BBa_K4274009) and degQ (Part: BBa_K4274010).
Usage and Biology
Our sfp gene (Gene ID: 45022253) is a biobrick part derived from B. amyloliquefaciens FZB42 encoding 4'-phosphopantetheinyl transferase. As mentioned above, the correct sfp gene is the core of lipopeptide biosurfactant production, thus the mutant sfp gene in B. subtilis is invalid for producing fengycins.
To construct a strain of B. subtilis to produce fengycins, we will use sfp_target (gRNA) (part: BBa_K4274008) to target the region of mutant sfp in B. subtilis for knock-out, simultaneously sfp (Part: BBa_K4274009) and degQ (Part: BBa_K4274010) gene were knocked-in in situ. It was used in the composite part PvanP*-sfp_target-sfp_HA_US-p43-K4274013-sfp-K4274014-degQ-sfp_HA_DS (Part: BBa_K4274035) to realize fengycins’ production in B. subtilis.
Characterization
The main composite of our product, fengycins, is produced by engineering Bacillus subtilis 168. As it is shown in the demonstration in description, sfp and degQ genes are critical to the biosynthesis of fengycins. Besides, Bacillus subtilis 168 possesses a natural but invalid sfp gene, and the functional degQ gene is silenced with the regulation of a weak promoter.Thus, with the help of pJOE8999_sfp_degQ plasmid (Figure 1a), we knocked in both sfp gene and degQ gene in the region of the natural sfp gene of Bacillus subtilis 168 after being knocking out. Under the guidance of protocol, we successfully transformed and induced the plasmid to function as a gene editor (Figure 1c). After induction, 7 strains were selected for PCR and electrophoresis verification. Since successful knock-in would would result in a 252 bps increase in the genome of Bacillus subtilis 168, the electrophoresis result conveys preliminarily that all of these strains has achieved our target of constructing the fengycins producers (Figure 1b).
After sequencing further verified that the successful construction of the fengycins producing strain, we used the strain for fermentation (Figure 2a). For the quantification of fengycins production, the methanol - extract obtained from the fermentation broth was analyzed by high performance liquid chromatography (HPLC). The peak of fengycins appeared at the retention time of 8-11.5 min under methanol mobile phase (Figure 2c). The injection sample was a solution of lipopeptide extract from 30 mL of fermentation broth dissolved in 7 mL methanol. Besides, various concentrations of fengycins standard is also injected for the construction of the standard curve of peak area obtained by HPLC (y) and its corresponding fengycins’ concentration (x). Based on the standard curve, we got the regression curve and its corresponding formula y=9.767x+193.44, and R2>0.98 displays that the regression equation fits the observed values (Figure 2b). Substituting y=1242.3 obtained from the sample into the regression equation, the concentration of fengycin in methanol solution can be calculated as 107.4 mg/L, that is, 25.06 mg/L of fengycins can be achieved by shaking-flask fermentation.
Source
B. amyloliquefaciens FZB42
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 38
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 38
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 38
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 38
Illegal NgoMIV site found at 95 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 661
References
[1]Chen X.H., Koumoutsi A., Scholz R., et al. More than Anticipated – Production of Antibiotics and Other Secondary Metabolites by Bacillus amyloliquefaciens FZB42. Mircrobial Biotech. 16 (2), 14-24 (2009). https://doi.org/10.1159/000142891.
[2]Jin P., Wang H., Liu W., et al. Characteriztion of IpaH2 gene corresponding to lipopeptide synthesis in Bacillus amyloliquefaciens HAB-2. BMC Microbio. 17 (2), 227 (2017). https://doi.org/10.1186/s12866-017-1134-z.
[3]Tsuge K., Ano T., Hirai M., et al. The Genes degQ, pps, and Ipa-8(sfp) Are Responsible for Conversion of Bacillus subtilis 168 to Plipastin Production. Antimicrobial Agents and Chemo. 43(9), 2183-2192 (1999). https://doi.org/10.1128/AAC.43.9.2183.
