Difference between revisions of "Part:BBa K185047"
Zhangyuanpu (Talk | contribs) (→Result) |
|||
| (23 intermediate revisions by 2 users not shown) | |||
| Line 15: | Line 15: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K185047 SequenceAndFeatures</partinfo> | <partinfo>BBa_K185047 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | == Characterized by CAFA_China 2022 == | ||
| + | *We constructed gene circuit contains UV-inducible switch “sulA promoter” and relE toxin. | ||
| + | |||
| + | [[File:SulA promoter and relE toxin.png|400px|thumb|center|Gene circuit: pSB1C3-sulAp-relE]]<br> | ||
| + | |||
| + | *We cultured the recombinant bacteria in the dark to optimum cell growth (OD600) and irradiated them under ultraviolet C. Then, each sample was cultured in incubating-shaker in darkness. | ||
| + | *The expression of relE gene results in bacterial growth slower than the control group, Showing the ability of relE to inhibit cell growth. | ||
| + | [[File:Inhibiting bacterial growth.png|400px|thumb|center|Figure: The OD600 of recombinant bacteria after irradiated under UVC (254nm). Competent host cell: DH10B. 0-11h: data collection time after UVC induction.]]<br> | ||
== Characterized by BNU-China 2019 == | == Characterized by BNU-China 2019 == | ||
| Line 65: | Line 75: | ||
This new information learned from literature that need attention was added by <html><a href="https://2020.igem.org/Team:Fudan" target="_blank">2020 Fudan team</a>.<br> | This new information learned from literature that need attention was added by <html><a href="https://2020.igem.org/Team:Fudan" target="_blank">2020 Fudan team</a>.<br> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| Line 76: | Line 83: | ||
[2] Jones, Brian V. “The human gut mobile metagenome: a metazoan perspective.” Gut microbes vol. 1,6 (2010): 415-31. doi:10.4161/gmic.1.6.14087.<br> | [2] Jones, Brian V. “The human gut mobile metagenome: a metazoan perspective.” Gut microbes vol. 1,6 (2010): 415-31. doi:10.4161/gmic.1.6.14087.<br> | ||
[3] Unterholzner, Simon J et al. “Toxin-antitoxin systems: Biology, identification, and application.” Mobile genetic elements vol. 3,5 (2013): e26219. doi:10.4161/mge.26219.<br> | [3] Unterholzner, Simon J et al. “Toxin-antitoxin systems: Biology, identification, and application.” Mobile genetic elements vol. 3,5 (2013): e26219. doi:10.4161/mge.26219.<br> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | == Characterized by HZAU_China 2023 == | ||
| + | *As a toxin protein, it does not have strong bactericidal effects, but has strong abilities to inhibit bacterial growth. We can use this toxin protein to reduce the influence of environmental temperature fluctuations on expression leakage of the RNA thermosensor. It can also play a role in inhibiting engineered bacterial growth in the early stage before the temperature-sensitive circuit activates engineered bacteria suicide. Its bacteriostatic effects are rapid and work well in the early stage according to our experiment. | ||
| + | ===Test Design=== | ||
| + | In our experiment, we constructed a plasmid vector with RelE toxin protein connected after the lactose promoter and introduced it into E.coli BL21(DE3). IPTG was used to induce expression and OD600 was detected continuously for 4 hours. Samples were taken every 30 minutes, diluted and plated to determine viable cell counts. | ||
| + | ===Test Protocol=== | ||
| + | <b>Toxin protein RelE OD600 detection</b> | ||
| + | <p> 1) Methods of molecular cloning and transformation are described above. Transform this plasmid into E. coli BL21. Then spread it onto LB medium plates with 50 μg/mL kanamycin and incubate overnight at 37 °C in an incubator. </p> | ||
| + | <p> 2) Pick four colonies from the same plate as parallel repeats. Each colony is inoculated in two identical media with 5 ml LB medium containing 50 μg/mL kanamycin and cultured at temperature (37 °C) respectively while shaking at 200 rpm. </p> | ||
| + | <p> 3) Measure the OD600 value of the resuspension culture media in an automatic microplate reader (SynergyH1 hybrid multimodal reader) until the OD600 of the bacteria solution reaches 0.4-0.6. </p> | ||
| + | <p> 4) Add IPTG to the bacteria solution of the experimental group to induce expression of the toxin proteins.</p> | ||
| + | <p> 5) Plot the OD600 value curves of the resuspension culture media over time in an automatic microplate reader (SynergyH1 hybrid multimodal reader). Incubate the cultures for 4 hours at 37°C while shaking at 200 rpm. Take samples at intervals or continuously measure the OD600 data of the bacteria solution with a UV spectrophotometer. And then convert the raw data into OD600. Compare the data of experimental groups and control group and compare curves in two schemes with each other.</p> | ||
| + | |||
| + | <b>Toxin protein RelE CFU detection</b> | ||
| + | <p> 1) 1. Methods of molecular cloning and transformation are described above. Transform this plasmid into E. coli BL21. Then spread it onto LB medium plates with 50 μg/mL kanamycin and incubate overnight at 37 °C in an incubator.</p> | ||
| + | <p> 2) Pick four colonies from the same plate as parallel repeats. Each colony is inoculated in two identical media with 5 ml LB medium containing 50 μg/mL kanamycin and cultured at temperature (37 °C) respectively while shaking at 200 rpm.</p> | ||
| + | <p> 3) Measure the OD600 value of the resuspension culture media in an automatic microplate reader (SynergyH1 hybrid multimodal reader) until the OD600 of the bacteria solution reaches 0.4-0.6. </p> | ||
| + | <p> 4) Take samples every half hour, dilute the bacteria solution 1*106times, then spread it onto LB medium plates with 50 μg/mL kanamycin and incubate overnight at 37 °C in an incubator.</p> | ||
| + | <p> 5)Count colonies on the plates cultured for the same time the next day, process the statistical data and plot the image.</p> | ||
| + | |||
| + | ===Result=== | ||
| + | <html> | ||
| + | <head> | ||
| + | <meta charset="utf-8"> | ||
| + | <title>无标题文档</title> | ||
| + | </head> | ||
| + | <body> | ||
| + | <center><img src="https://static.igem.wiki/teams/4645/wiki/wet-lab/suicide/rele2-1.jpg" style="width:50%; "></center> | ||
| + | <br> | ||
| + | <center><b>Figure 1. Bacterial OD600 over time .</b> </center> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <center><img src="https://static.igem.wiki/teams/4645/wiki/wet-lab/suicide/rele2-2-ab.jpg" style="width:80%; "></center> | ||
| + | <br> | ||
| + | <center><b>Figure 2. Viable cell counts over time.</b></center> | ||
| + | <br> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | ===Analysis of the experimental results=== | ||
| + | Since RelE is a weak toxin protein, as shown in Figure 1., the OD600 detection showed obvious inhibition of bacterial growth after induced expression, which delayed the log phase, but the bacteria could still grow in the later phase. The viable cell count method also produced significant results. As shown in Figure 2., viable cell counts decreased rapidly within one hour after induction, exhibiting significant growth inhibition, which demonstrated the bacteriostatic effect of this toxin. | ||
Latest revision as of 01:16, 11 October 2023
RelE toxin
The relE toxin is an RNase that preferentially cleaves mRNAs bound to the ribosome at the second position of stop codons. Stop codons not only signal the end of the protein coding sequence but also serve as the binding site for release factors, which promote release of the nascent polypeptide and facilitate recycling of ribosomes for further rounds of translation. Thus truncated mRNA by cleavage of relE lacks appropriate termination signals, which causes the accumulation of stalled ribosomes and these mRNAs are unable to promote release factor binding, nascent polypeptide release, and ribosome recycling. As a result, expression of the relE gene has been shown to severely inhibit translation and prevent colony formation. RelE display codon-specific cleavage of mRNAs in the ribosomal A site, that is to say, among stop codons UAG is cleaved with fast, UAA intermediate and UGA slow rate(UAG>UAA>UGA).
In our design, we add a his-tag at the end of RelE sequence to detect the expression of relE protein. Then we mutated the stop condon from UGA to UAA to make relE toxin inhibit its own traslation moderately.Most importantly, you can get this part in BBa_K185000 or BBa_K185004
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterized by CAFA_China 2022
- We constructed gene circuit contains UV-inducible switch “sulA promoter” and relE toxin.
- We cultured the recombinant bacteria in the dark to optimum cell growth (OD600) and irradiated them under ultraviolet C. Then, each sample was cultured in incubating-shaker in darkness.
- The expression of relE gene results in bacterial growth slower than the control group, Showing the ability of relE to inhibit cell growth.
Characterized by BNU-China 2019
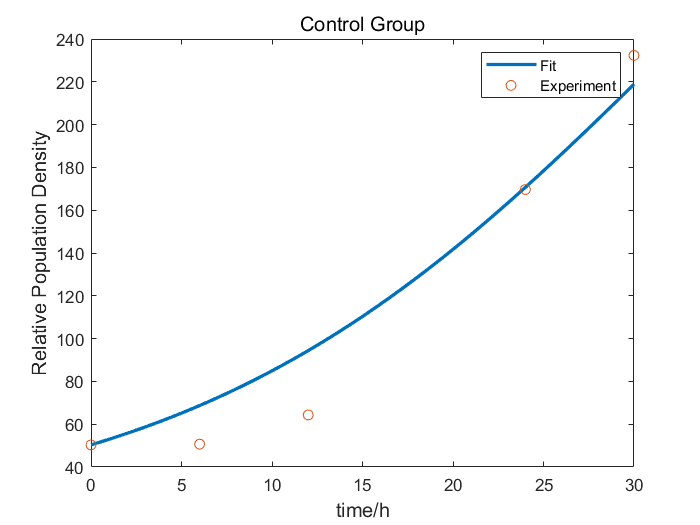
We characterize relE (BBa_K185000) by an antitoxin-toxin system, in which the downstream relE gene encoding for a stable toxin is constantly expressed, and the upstream relB (BBa_K185048) gene encodes for a labile antitoxin under the control of a temperature-sensitive RNA thermometer (BBa_K115002). Without counteracted by antitoxin RelB, RelE shuts down protein synthesis and causes the death of microbe by cleaving mRNA within the ribosomal A site [1]. In addition, the RNA thermometer allows expression of RelB at 37℃, but inhibits expression at 27℃, which leads to excess expression of RelE. As a result, we can characterize relE in a cell density-dependent manner in Escherichia coli K-12.
In order to characterize the toxicity of protein encoded by relE and whether it can be inhibited by antitoxin RelB, we take E. coli introduced with a vector with the same backbone as control group.
As is shown in Fig.1, there is nearly no difference of the relative population density between control and experimental groups at 37℃, which indicates RelE interacts with RelB and thereby the cleavage of mRNA by RelE is inhibited. However, the population density of experimental group shows a significant decrease compared to control group at 27℃, which indicates the protein encoded by relE is lethal to E. coli.
With properties of relE, we can construct a kill switch in engineered microbe to ensure lab safety.
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells. 2. A strain containing a vector with same backbone is used as control. Experimental groups and control groups are both cultured in 60mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm; 3. Equally divide each group into two flasks, which is 30mL respectively. One of each group is cultured at 27℃, 200rpm and the other at 37℃, 200rpm; 4. Extract 5μl samples of each culture system every 6 hours. Diluted all of the samples to 107 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately; 5. Count the number of colonies in 5 cm2 per plate after cultured for 24 hours at 37℃ 6. Three repicas are tested in each group.
Model (Added by 2020 Fudan)
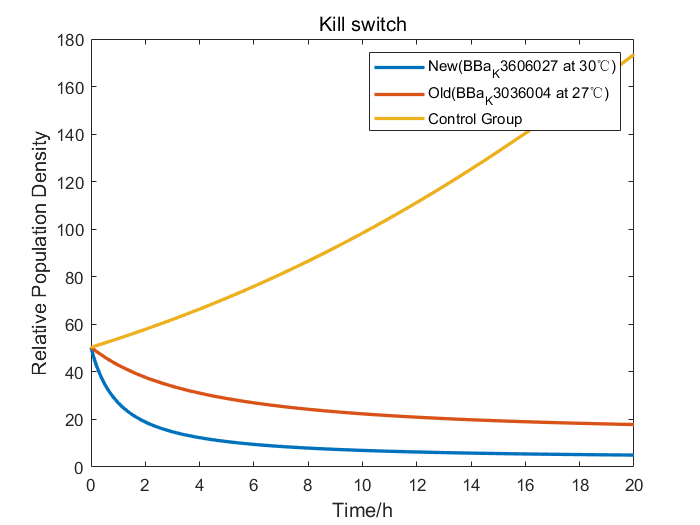
We collaborate with BNU-China and adopt their raw wet lab experimental data of RelE/RelB system to construct our model. After studying the data from BNU-China, we found out that without Kill Switch, the colony number curve of control group was fairly similar to the logistic equation. So, we fitted the experimental data to obtain the parameters of the logistic equation. When the Kill Switch exists, the toxin sharply increased the competitive pressure, introducing a parameter of additional pressure α into the equation. Therefore, we fitted the data of cold shock (27℃) group to figure out the value of α.
Furthermore, we performed an efficiency analysis by the adjusted model we had constructed. Our design of BBa_K3606027 employed a more efficient RNA thermometer than that of BNU-China, so we changed the properties to simulate the growth curve of our Kill Switch. The result demonstrated that bacteria carrying our Kill Switch were killed more thoroughly in a smaller temperature range, therefore proved that with our Kill Switch, engineered bacteria would have less deleterious effects on the environment.
Attention (Added by 2020 Fudan)
Potential pathogenicity of RelE
Although, RelE/RelB module poses minimized adverse effects on hosts as it expanding in microbial communities [2], it has several other effects on human intestine. Firstly, RelE is a very rare toxin that has the activity against both prokaryotic and eukaryotic organisms in a similar mechanism. It has been observed that RelE can induce apoptosis in cultured human cell lines. Secondly, RelE/RelB module tends to have unexpected enrichment in bacteria that is not observed for other toxin/antitoxin modules such as MazE/MazF and ParD/ParE. Such enrichment may be associated with the formation of persistence of microbes in the gut. Thirdly, the divisions of bacteria carrying homologues of the RelE toxin involved in comprising the human gut microbiota.
All this effect only been testified in artificial mammalian gene expression systems, so whether it has tangible impact on the human gut microbiome need further study. iGEM teams that focus on intestinal probiotics and have special safety requirements ought to pay special attention to these.
Chromosomally encoded RelE/RelB in gut microbiota
What’s more, RelE/RelB and other toxin/antitoxin modules have homology searches on bacterial chromosomes. RelB-RelE complex can inhibit the promotor of chromosomally encoded RelE/RelB [3].
This new information learned from literature that need attention was added by 2020 Fudan team.
Reference
[1] Overgaard M , Borch J , Gerdes K . RelB and RelE of Escherichia coli Form a Tight Complex That Represses Transcription via the Ribbon–Helix–Helix Motif in RelB[J]. Journal of Molecular Biology, 2009, 394(2):0-196.
[2] Jones, Brian V. “The human gut mobile metagenome: a metazoan perspective.” Gut microbes vol. 1,6 (2010): 415-31. doi:10.4161/gmic.1.6.14087.
[3] Unterholzner, Simon J et al. “Toxin-antitoxin systems: Biology, identification, and application.” Mobile genetic elements vol. 3,5 (2013): e26219. doi:10.4161/mge.26219.