Difference between revisions of "Part:BBa K4439007"
Charlottedml (Talk | contribs) |
Charlottedml (Talk | contribs) |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
==Abstract== | ==Abstract== | ||
| − | + | For the scope of our project, we designed a new recombinant silk protein which adds hydrophobic protective properties to the insulative cellulose aerogel we produced, when coated on top of it. On one hand, by fusing the protein sequence of the green lacewing silk to a Cellulose Binding Domain (CBD) on the C-terminus, the recombinant protein is able to bind the cellulose aerogel. On the other hand, by adding a streptavidin monomer (mSA) to the N-terminus, we can link to this silk fusion protein any other protein with interesting properties via the well known biotin-streptavidin interaction. Finally, to purify the resulting protein from BL21(DE3) E. Coli competent cells, the expression system we chose, we added a 10xHis tag to the C-terminus, after the CBD. | |
==Sequence and Features== | ==Sequence and Features== | ||
<partinfo>BBa_K4439007 SequenceAndFeatures</partinfo> | <partinfo>BBa_K4439007 SequenceAndFeatures</partinfo> | ||
| − | ==Protein Characterization== | + | ==Protein Characterization - Usage and Biology== |
| − | === | + | ====mSA==== |
| + | mSA, or monomeric streptavidin, is an engineered streptavidin/rhizavidin hybrid that binds to biotin with high affinity as a monomer, it was constructed and characterized by Lim et al. in 2013. It can be fused as a genetic tag to heterologous proteins to enable biotin binding. The biotin affinity of mSA is the highest among nontetrameric streptavidin, the monomer also has significantly higher stability and solubility than all other previously engineered monomers to ensure the folding and functionality of the molecule during its application. It is therefore a useful tool for biotechnology applications making use of the streptavidin-biotin interaction. | ||
| − | + | ====Recombinant Silk Protein==== | |
| − | + | Silk proteins demonstrate interesting mechanical properties such as toughness, strength, lightweight, biodegradability and the possibility to produce different morphologies (fibers, foams, capsules, films). In addition to this, silk proteins comprise a high percentage of the amino acids glycine, serine and alanine which have an intermediate hydrophobicity. | |
| − | + | Green lacewing insects produce two types of silk: one produced by the larvae (cocoon) and the other by adult females (egg-stalk). The adult produced silk acts as a protective shelter and structural support for egg stalks, which are two ideal properties for a waterproof coating for our aerogel. | |
| − | + | In green lacewings, two serine- and glycine-rich silk proteins (Ma1XB1 and Ma1XB2) have been identified, both with highly repetitive core domains and small terminal domains. The core domain’s structure is rich in β-sheets with an approximative sheet-length of four amino acids between turns. These form repeating structural units constituting β-helices which have a significant positive correlation with the proteins’ surface hydrophobicity. A consensus motif for the core domain of Ma1XB2 (named [AS])had already been generated. Furthermore, a recombinant protein constituted by 8 repetitions of this [AS] module had also already been expressed in E. coli. | |
| + | We decided to use the N[AS]4C protein in our plasmid construct in the end because the gene synthesis of 8 repetitive elements is quite difficult to perform. Also, this allowed us to have a smaller protein in the end, which leads to less risks of problems in the expression. | ||
| + | |||
| + | ====Cellulose-Binding Domain==== | ||
| + | |||
| + | The Imperial 2014 iGEM team ([[Part:BBa K1321014]]) documented a particular cellulose-binding domain (CBDCipA). The Puiching Macau 2020 iGEM team ([[Part:BBa K3503004]]) used their records to fuse the CBDCipA to the SR protein, another protein that we also used in our project. In addition, the Linkoping Sweden 2019 iGEM team ([[Part:BBa K3182001]]) pursued the characterisation of CBDCipA. As this particular cellulose binding domain originates from a thermophilic bacteria (which further increases the domain's applications) and was well documented on the iGEM Parts Registry, we decided to build upon the work of those three iGEM teams. | ||
| + | |||
| + | For the scope of our project, we reused the useful composite part CBD-SR from PuiChing Macau 2020 as it was and implemented their CBD sequence in our designs. | ||
===Modeling=== | ===Modeling=== | ||
| + | ====Protein Modeling==== | ||
<br> | <br> | ||
[[File:Model 01a.jpeg]] | [[File:Model 01a.jpeg]] | ||
<br> | <br> | ||
| − | Figure | + | Figure 1 | AlphaFold2 prediction for 01a. (A) First rank 3D model prediction of 01a protein; the iteration who got the highest score in the modeling. (B) Different correlation graphs between the query sequence of 01a and the predicted one, for each proposed 5 models. (C) Figure of sequence coverage of 01a and indices on alignment with other sequences in the mSA. (D) IDDT graph for 01a per residue to get an idea of the confidence of the model in predicting the geometry. |
| − | * Analysis : According to (fig. | + | * Analysis : According to (fig. 1, A), we could identify the structure of the different single chains of interest easily. The blue slightly smaller helicoidal structure represents the mSA protein that would be attached to the silk protein, the elongated green structure and finally we would get the CBD sequence that is displayed in red. The (fig.1, B) confirms the hypothesis of knowing very efficiently the three domains and having trouble distinguishing the in-between linkage which is totally normal since we designed those linking segments to be able to keep the structure of the three main elements. The same graph showed that few iterations of the prediction lacked to characterize the silk segment, which could be an issue in the experiments. However both (fig. 1, C) and (fig. 1, D), confirmed the reasoning that our proteins would keep a structure preserving their initial aim. The linkage would give in reality more freedom of placement of those chains. The IDDT score helped to identify the percentage of correctly predicted and true structure that could be superimposed. In general, the higher this score the better the model is considered. |
| + | ====Modeling the amount of proteins to coat a cellulose aerogel==== | ||
| − | + | We had different parameters that needed to be considered and we listed the most of them in (Table. 1) | |
<br> | <br> | ||
| − | [[File: | + | [[File:Aerogel table value.png|500px]] |
<br> | <br> | ||
| − | Figure 1 | Protein Purification Protocol using Ni-NTA Beads | + | <br> |
| + | [[File:Protein table value.png|500px]] | ||
| + | <br> | ||
| + | Table 1: Table for the parametric model . | ||
| + | |||
| + | In the proposed model we would like to coat geometrically our aerogel surface with different CBD globular entities. Since they are the most probable point of anchoring they will represent an entire protein in the counting of the amount of proteins. We supposed that the proteins would be placed in an adjacent manner. | ||
| + | |||
| + | First step was to consider the aerogel as a perfect cylinder that will be coated by the proteins. The surface of the cylinder is simplified to two circles and an elongated rectangle (fig2). We proceeded to a geometrical filling of the surface. | ||
| + | |||
| + | <br> | ||
| + | [[File:Aerogel Cylinder Plots.jpeg]] | ||
| + | <br> | ||
| + | Figure 2 | Aerogel simplified representation as a perfect cylinder. | ||
| + | |||
| + | The attaching CBD region was taken from the above model 4JO5 to study its hydrodynamic characteristics using the Hullrad algorithm. This program was used to get specific useful information on the protein 4JO5: | ||
| + | |||
| + | <br> | ||
| + | [[File:Table values modeling proteins.png|500px]] | ||
| + | <br> | ||
| + | Table 2 | Hullrad computation for CBD, AJO5 | ||
| + | |||
| + | Using the values from (Table. 2), we populated the different surfaces of the aerogel with circular proteins using some simple geometry equations. The two values of AVSR and MD were studied in the example. Considering the protein radius and aerogel diameter, we came up with an iterative algorithm in three steps: | ||
| + | Step 1: Populate the outer peripheral of the bigger cylinder: | ||
| + | |||
| + | We used the following angular equation to compute the angle of placement of the first small protein circle in the aerogel bigger circle: | ||
| + | |||
| + | alpha = 1 /π |arcsin(r(proteins)/(r(aerogel)- r(proteins)))*180| (1) | ||
| + | |||
| + | where r(proteins) and r(aerogel) represent respectively the radius of the proteins and the aerogel. | ||
| + | |||
| + | Step2: Compute the number of cylinders that can populate the peripheral of our the bigger circle: | ||
| + | |||
| + | nb(circles) =[360/(2*alpha)] (2) | ||
| + | where [x] is the greatest integer function s. | ||
| + | |||
| + | |||
| + | Step 3: Define a secondary circle that has the next small circles: | ||
| + | |||
| + | We iterated the new radius until we could not fit any small circles in the area (fig.12): | ||
| + | |||
| + | r(new)=r(previous)-( 2 *r(small)) (3) | ||
| + | where r(previous) is the large radius of the previous iteration. | ||
| + | |||
| + | Ending condition : | ||
| + | r(previous)< r(small) | ||
| + | |||
| + | <br> | ||
| + | [[File:Populated cylinder aerogel.jpeg]] | ||
| + | <br> | ||
| + | Figure 3 | Populating circles with tiny cylinders zoom at 150 Å aerogel surface . | ||
| + | |||
| + | In (fig.3), we saw clearly how the different circles were placed inside the allocated surface of aerogel. We extended the reasoning to filling the rectangular lateral surface. | ||
| + | |||
| + | For the lateral specific surface, the operation is described as following: | ||
| + | |||
| + | Step 1: Divide the width of the rectangle by the number of small protein circles that fits in the line : | ||
| + | |||
| + | nb(circles for width) = [W2 * r(proteins)] (4) | ||
| + | where W is the width of the rectangle, r(proteins) represents the radius of the proteins and [x] is the greatest integer function as described above. | ||
| + | |||
| + | Step 2: Multiply the above number by the number of lines that can fit in the height of | ||
| + | the rectangle: | ||
| + | |||
| + | nb(circles for rectangle) = [h2* r(small)] * nb(circles for length) (5) | ||
| + | where h represents the height of the rectangle, [x] is the greatest integer function as described above. | ||
| + | |||
| + | By taking the parameters of the aerogel from (Table.1) and the parameters of the CBD from (Table.2) we got the following output in number of molecules and in number of molar: | ||
| + | |||
| + | <br> | ||
| + | [[File:Table values.png|500px]] | ||
| + | <br> | ||
| + | Table 3: Results of the coating computations. | ||
| + | |||
| + | *The values found in (Table.3) were useful to define the protein solution volume that we soaked the hydrogels or the aerogels with. | ||
| + | |||
| + | ==Silk Specific Experiments Processes== | ||
| + | |||
| + | <br> | ||
| + | [[File:His-tagged Protein Purification.png|600px]] | ||
| + | <br> | ||
| + | Figure 4 | Protein Purification Protocol using Ni-NTA Beads | ||
*For more information, check out our Protocols page on our wiki. | *For more information, check out our Protocols page on our wiki. | ||
| Line 43: | Line 133: | ||
[[File:Silk_Biofilm_EPFL.png|500px]] | [[File:Silk_Biofilm_EPFL.png|500px]] | ||
<br> | <br> | ||
| − | Figure | + | Figure 5 | Diagram of Procedure for Silk Biofilm Fabrication |
*For more information, check out our Protocols page on our wiki. | *For more information, check out our Protocols page on our wiki. | ||
| Line 52: | Line 142: | ||
*Purification might prove difficult : elution with EDTA works better but might damage the protein more. | *Purification might prove difficult : elution with EDTA works better but might damage the protein more. | ||
| − | *The silk biofilm can be created even if the recombinant protein is in liquid form. | + | *The silk biofilm can be created even if the recombinant protein is in liquid form. |
| + | |||
| + | *When using formic acid, follow carefully the safety guidelines indicated on the Material Safety Datasheet. | ||
==Results== | ==Results== | ||
| Line 63: | Line 155: | ||
[[File:Transfo 01a BL21 EPFL.jpeg]] | [[File:Transfo 01a BL21 EPFL.jpeg]] | ||
<br> | <br> | ||
| − | Figure | + | Figure 6 | Bacterial transformation with the 01a construct in BL21(DE3) competent cells. (A) The plate transformed with 01a. (B) The plate transformed with a plasmid containing standard GFP (positive control). (C) The plate transformed with water (negative control). |
After the overnight incubation, we observed the presence of colonies in the BL21(DE3) transformed cells plates of 01a and the positive control, no colonies were observed on the negative control, as expected. | After the overnight incubation, we observed the presence of colonies in the BL21(DE3) transformed cells plates of 01a and the positive control, no colonies were observed on the negative control, as expected. | ||
| Line 71: | Line 163: | ||
The 01a GeneScript plasmid transformation worked well for BL21(DE3) since we can observe colonies on the LB-Kanamycin plates and the OD600 of the transformation liquid culture is high, meaning that many bacteria were able to grow. | The 01a GeneScript plasmid transformation worked well for BL21(DE3) since we can observe colonies on the LB-Kanamycin plates and the OD600 of the transformation liquid culture is high, meaning that many bacteria were able to grow. | ||
| − | ===Purification=== | + | ===Protein Purification=== |
The purification was performed following a Ni-NTA beads based protocol. | The purification was performed following a Ni-NTA beads based protocol. | ||
| Line 77: | Line 169: | ||
[[File:Purification results 01a.png|900px]] | [[File:Purification results 01a.png|900px]] | ||
<br> | <br> | ||
| − | Figure | + | Figure 7 | Protein purification of silk fusion protein (01a)(A) SDS-PAGE gel stained with Coomassie Blue Protein Stain of all the fractions of silk fusion protein purification (B) Western Blot of the elution 1 (E1 fraction on SDS-PAGE gel) visualised with anti-His antibody. W1 = Wash 1 with 20 mM imidazole; W2 = Wash 2 with 50 mM imidazole; E1 = Elution 1 with 250 mM imidazole; E2 = Elution 2 with 500 mM imidazole; E4 = 2.5 M imidazole; E5 = 5 M imidazole. |
*Analysis | *Analysis | ||
| − | Protein purification of the mSA-N[AS]4C-CBD-10xHis protein (01a) showed a 100kDa protein present in the first elution lane (E1) in both the SDS-PAGE (fig | + | Protein purification of the mSA-N[AS]4C-CBD-10xHis protein (01a) showed a 100kDa protein present in the first elution lane (E1) in both the SDS-PAGE (fig 7.A) and the Western Blot (fig 7.B). mSA-N[AS]4C-CBD-10xHis protein was expected at 78kDa size. Using the Nanodrop, we measured the concentration (0.72 mg/mL) and we obtained a final amount of 2.15 mg of the mSA-N[AS]4C-CBD-10xHis protein (01a). |
Since high imidazole concentration might denature the proteins, we had to either remove it or make imidazole inert. It was not possible to dialyse the proteins to remove imidazole, since the dialysis membrane was made of cellulose, and our fusion proteins would all bind to the membrane via their cellulose binding domain (CBD). We therefore decided to flash freeze the proteins and store them in the elution buffers so the imidazole will no longer affect them when frozen. | Since high imidazole concentration might denature the proteins, we had to either remove it or make imidazole inert. It was not possible to dialyse the proteins to remove imidazole, since the dialysis membrane was made of cellulose, and our fusion proteins would all bind to the membrane via their cellulose binding domain (CBD). We therefore decided to flash freeze the proteins and store them in the elution buffers so the imidazole will no longer affect them when frozen. | ||
| − | === | + | ===Cloning by digestion-ligation=== |
| + | The elution of the proteins of interest in such high amounts of imidazole certainly comes from the double His-tag that composes our proteins. To remove it from the received plasmids, we performed some cloning experiments to obtain better yields in the future purifications. In the case of the silk fusion protein, because of its repetitive modules the standard PCR-KLD cloning. To remove the added part, we therefore digested our plasmid (fig 8.A) with the NcoI enzyme since there are two NcoI restriction sites on both sides of the added site (fig 8.B) and re-ligated it back (fig 8.C). | ||
| + | <br> | ||
| + | [[File:Digestion Ligation 01a.png|900px]] | ||
| + | <br> | ||
| + | Figure 8 | Cloning experiment results for the removal of the added site for 01a (silk). (A) Agarose gel electrophoresis of the digested product of 01a. (B) Plasmid map of 01a (silk) with the added part. (C) Plasmid map from the sequencing result of the obtained new 01a plasmid (silk). | ||
| + | |||
| + | <br> | ||
| + | [[File:Purification trial4 01a.png|900px]] | ||
| + | <br> | ||
| + | Figure 9 | Expression of the silk fusion protein from the plasmid without the added site. (A) SDS-PAGE gel stained with Coomassie Blue Protein Stain of the lysate before and after IPTG induction. (B) SDS-PAGE gel stained with Coomassie Blue Protein Stain of the samples from protein purification of the new silk fusion protein induced in BL21(DE3). SN = Supernatant; FT = Flowthrough; W1 = Wash 1 with 20 mM imidazole; W2 = Wash 2 with 50 mM imidazole; E1 = Elution 1 with 250 mM imidazole; E2 = Elution 2 with 500 mM imidazole. | ||
| + | |||
| + | *Analysis | ||
| + | Since we obtained the wanted plasmid, we transformed new BL21(DE3) competent cells to start a new protein production. By doing a gel comparing the protein expression before and after IPTG induction (fig 9.A), we could see a band at approximately 100 kDa. Protein purification of the silk fusion protein (01a) also showed a 100kDa protein present in the two elution lanes (E1 and E2). Even though we expected the protein to be around 74kDa, this 100kDa lane should correspond to our new silk fusion protein, since we obtained similar ones in the previous purification. By measuring the concentration (0.37 mg/mL), we obtained a final amount of 11 mg of the silk fusion protein (01a) | ||
| + | |||
| + | === Biofilm Fabrication=== | ||
<br> | <br> | ||
[[File:Silk Biofilm 01a.jpeg]] | [[File:Silk Biofilm 01a.jpeg]] | ||
<br> | <br> | ||
| − | Figure | + | Figure 10 | Silk biofilms inside polystyrene petri dishes. (A) Silk fusion protein biofilm in polystyrene petri dish after 24 hours drying. (B) GFP fusion protein biofilm in polystyrene petri dish after 24 hours drying. |
*Analysis | *Analysis | ||
| − | After the production of our proteins, we created a silk fusion protein biofilm to coat an aerogel to make it water-resistant. Inspired by the work of Felix Bauer, we produced two biofilms: one with the recombinant silk protein (fig | + | After the production of our proteins, we created a silk fusion protein biofilm to coat an aerogel to make it water-resistant. Inspired by the work of Felix Bauer, we produced two biofilms: one with the recombinant silk protein (fig 10.A) and one with the recombinant GFP protein, as a control (fig 10.B). In figure 8, we can see that we dried the biofilms too much due to the presence of cracks. Even if it was not a problem for hydrophobicity testing, it became one to remove the biofilm from the dish. Indeed, it was impossible for us to take the biofilm off, even by cutting the edges of the petri dish. This meant that we couldn’t coat aerogels with those biofilms. However, we saw that the GFP biofilm (fig 10.B) made some crystals and cubic structures so that the intended biofilm was less homogeneous than the silk one (fig 10.A). Overall, we managed to generate a biofilm with our recombinant silk fusion protein (mSA-silk-CBD), but not with the GFP fusion protein (mSA-GFP-CBD). |
| − | ===Hydrophobicity | + | ===Hydrophobicity Test=== |
| − | To | + | With these biofilms, we did a wettability test to compare their hydrophobicity. To do so, we added droplets of water on top of those biofilms and did several measurements. |
| + | |||
| + | <br> | ||
| + | [[File:Hydrophobicity Test 01a.jpeg]] | ||
| + | <br> | ||
| + | Figure 11 | Silk biofilms with water droplets for hydrophobicity tests. (A) Silk fusion protein biofilm (01a) with a drop of water (B) GFP fusion protein biofilm (01a) with a drop of water. (C) Measurement of the angle made by the drop on the silk biofilm. (D) Measurement of the angle made by the drop on the GFP biofilm. (E) Dried drops of water on the silk biofilm (F) Dried drops of water on the GFP biofilm. | ||
| + | |||
| + | *Analysis | ||
| + | First, we measured the angle made by the droplet (fig 11.A and B) to assess surface tension and the surface wettability. Then we measured the time it took for the water to be absorbed or evaporated, computed the mean values for each biofilm and compared them to conclude about hydrophobicity. For the first measures, we obtained an angle of 55.2° for the silk (fig 11.C) and 27.3° for the GFP biofilm (fig 11.D). This meant that the silk fusion protein biofilm (01a) had a higher contact angle than the GFP biofilm. Moreover, by measuring the time it takes for three water droplets to be absorbed, we obtained a mean of 466,33 s for the silk biofilm (fig 11.E), and 28,33 s for the GFP biofilm (fig 11.F). With these values, we concluded that it takes more time for a drop of water to be evaporated or absorbed from the silk biofilm. These two results suggested that our biofilm helps protect the aerogel from water droplets. | ||
| + | |||
| + | ===Coating an aerogel with mSA-N[AS]4C-CBD-10xHis protein=== | ||
| + | We showed that coating the aerogel with the silk fusion protein makes it more water resistant than without the coating. We first coated hydrogels with proteins (either the silk fusion protein (01a), the GFP fusion protein (03a), the GFP control protein or water) (fig 12.A) and sent them to freeze dry (fig 12.B). After this step, we saw that the coated aerogels had a more homogeneous shape, with less cracks and a sort of protective film on top of it compared to the uncoated ones (fig 13.A). | ||
| + | |||
| + | <br> | ||
| + | [[File:Aerogels Hydrogels set-up 01a.png|700px]] | ||
| + | <br> | ||
| + | Figure 12 | Freeze drying of coated hydrogels. (A) Hydrogels before freeze drying. (B) Aerogel after 23 hours of freeze drying. | ||
| + | |||
| + | <br> | ||
| + | [[File:Coating silk.jpeg]] | ||
| + | <br> | ||
| + | Figure 13 | Protective coating given by the silk fusion protein. (A) Comparison between uncoated and coated with recombinant silk (01a) aerogels. (B) Boxplots of the delay time a drop of water gets absorbed inside an aerogel coated with the different proteins. Top of the boxes = mean of delay time per group; Black dots = individual delay time measurements; Red vertical line = standard error of the mean. Data are mean ± s.d., n = 3 measurements per group; ***P < 0.001, **P < 0.01; one way ANOVA test was performed followed by a Tukey HSD for performing multiple pairwise-comparisons between the means of groups. | ||
| + | |||
| + | *Analysis | ||
| + | Using the Keyence microscope, we filmed the application of 4 to 5 drops of water on the aerogels and quantified the water absorption time. For each aerogel we computed the mean delay time and used this data to perform a one-way ANOVA test to see if there is a significant difference between the means between the groups. We obtained a final p-value of 0.000576. As the p-value was less than the significance level 0.05, we concluded that there are significant differences between the groups. However, this result did not indicate which pairs of groups are different. As the ANOVA test was significant, we computed a Tukey HSD for performing multiple pairwise-comparisons between the means of groups. For each comparison with the silk fusion protein (01a), we obtained a p-value way smaller than 0.05, which means that there was a significant difference between the silk fusion protein and the other groups, while there was no significant difference among the other groups (dH20, GFP and GFP fusion protein 03a). Therefore we concluded that aerogels absorb water more slowly when they are coated with the silk fusion protein. Indeed, in the box plot we plotted to present the different populations (fig 13.B), it is clear that in the case of the silk fusion protein (01a), the delay times were higher. The water drop took longer to be fully absorbed by the aerogel coated with the recombinant silk, suggesting that the recombinant silk adds hydrophobicity to the aerogel. Overall, these results proved that the CBD domain we added to our silk construct allows binding to the aerogel and that this protein provides a protective coating for the aerogel. | ||
==References== | ==References== | ||
| + | |||
| + | <ul> | ||
| + | <li> Bauer, F. & Scheibel, T. (2012, 16 mai). Artificial Egg Stalks Made of a Recombinantly Produced Lacewing Silk Protein. Angewandte Chemie International Edition, 51(26), 6521‑6524. https://doi.org/10.1002/anie.201200591 | ||
| + | |||
| + | <li> Bauer, F. (2013) Development of an artificial silk protein on the basis of a lacewing egg stalk protein. https://epub.uni-bayreuth.de/72/1/Diss.pdf | ||
| + | |||
| + | <li> Lim, K. H., Huang, H., Pralle, A. & Park, S. (2012, août 8). Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnology and Bioengineering, 110(1), 57‑67. https://doi.org/10.1002/bit.24605 | ||
| + | |||
| + | |||
| + | </li> | ||
| + | </ul> | ||
Latest revision as of 15:17, 12 October 2022
mSA-N[AS]4C-CBD-10xHis
Contents
Abstract
For the scope of our project, we designed a new recombinant silk protein which adds hydrophobic protective properties to the insulative cellulose aerogel we produced, when coated on top of it. On one hand, by fusing the protein sequence of the green lacewing silk to a Cellulose Binding Domain (CBD) on the C-terminus, the recombinant protein is able to bind the cellulose aerogel. On the other hand, by adding a streptavidin monomer (mSA) to the N-terminus, we can link to this silk fusion protein any other protein with interesting properties via the well known biotin-streptavidin interaction. Finally, to purify the resulting protein from BL21(DE3) E. Coli competent cells, the expression system we chose, we added a 10xHis tag to the C-terminus, after the CBD.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Protein Characterization - Usage and Biology
mSA
mSA, or monomeric streptavidin, is an engineered streptavidin/rhizavidin hybrid that binds to biotin with high affinity as a monomer, it was constructed and characterized by Lim et al. in 2013. It can be fused as a genetic tag to heterologous proteins to enable biotin binding. The biotin affinity of mSA is the highest among nontetrameric streptavidin, the monomer also has significantly higher stability and solubility than all other previously engineered monomers to ensure the folding and functionality of the molecule during its application. It is therefore a useful tool for biotechnology applications making use of the streptavidin-biotin interaction.
Recombinant Silk Protein
Silk proteins demonstrate interesting mechanical properties such as toughness, strength, lightweight, biodegradability and the possibility to produce different morphologies (fibers, foams, capsules, films). In addition to this, silk proteins comprise a high percentage of the amino acids glycine, serine and alanine which have an intermediate hydrophobicity.
Green lacewing insects produce two types of silk: one produced by the larvae (cocoon) and the other by adult females (egg-stalk). The adult produced silk acts as a protective shelter and structural support for egg stalks, which are two ideal properties for a waterproof coating for our aerogel.
In green lacewings, two serine- and glycine-rich silk proteins (Ma1XB1 and Ma1XB2) have been identified, both with highly repetitive core domains and small terminal domains. The core domain’s structure is rich in β-sheets with an approximative sheet-length of four amino acids between turns. These form repeating structural units constituting β-helices which have a significant positive correlation with the proteins’ surface hydrophobicity. A consensus motif for the core domain of Ma1XB2 (named [AS])had already been generated. Furthermore, a recombinant protein constituted by 8 repetitions of this [AS] module had also already been expressed in E. coli.
We decided to use the N[AS]4C protein in our plasmid construct in the end because the gene synthesis of 8 repetitive elements is quite difficult to perform. Also, this allowed us to have a smaller protein in the end, which leads to less risks of problems in the expression.
Cellulose-Binding Domain
The Imperial 2014 iGEM team (Part:BBa K1321014) documented a particular cellulose-binding domain (CBDCipA). The Puiching Macau 2020 iGEM team (Part:BBa K3503004) used their records to fuse the CBDCipA to the SR protein, another protein that we also used in our project. In addition, the Linkoping Sweden 2019 iGEM team (Part:BBa K3182001) pursued the characterisation of CBDCipA. As this particular cellulose binding domain originates from a thermophilic bacteria (which further increases the domain's applications) and was well documented on the iGEM Parts Registry, we decided to build upon the work of those three iGEM teams.
For the scope of our project, we reused the useful composite part CBD-SR from PuiChing Macau 2020 as it was and implemented their CBD sequence in our designs.
Modeling
Protein Modeling

Figure 1 | AlphaFold2 prediction for 01a. (A) First rank 3D model prediction of 01a protein; the iteration who got the highest score in the modeling. (B) Different correlation graphs between the query sequence of 01a and the predicted one, for each proposed 5 models. (C) Figure of sequence coverage of 01a and indices on alignment with other sequences in the mSA. (D) IDDT graph for 01a per residue to get an idea of the confidence of the model in predicting the geometry.
- Analysis : According to (fig. 1, A), we could identify the structure of the different single chains of interest easily. The blue slightly smaller helicoidal structure represents the mSA protein that would be attached to the silk protein, the elongated green structure and finally we would get the CBD sequence that is displayed in red. The (fig.1, B) confirms the hypothesis of knowing very efficiently the three domains and having trouble distinguishing the in-between linkage which is totally normal since we designed those linking segments to be able to keep the structure of the three main elements. The same graph showed that few iterations of the prediction lacked to characterize the silk segment, which could be an issue in the experiments. However both (fig. 1, C) and (fig. 1, D), confirmed the reasoning that our proteins would keep a structure preserving their initial aim. The linkage would give in reality more freedom of placement of those chains. The IDDT score helped to identify the percentage of correctly predicted and true structure that could be superimposed. In general, the higher this score the better the model is considered.
Modeling the amount of proteins to coat a cellulose aerogel
We had different parameters that needed to be considered and we listed the most of them in (Table. 1)

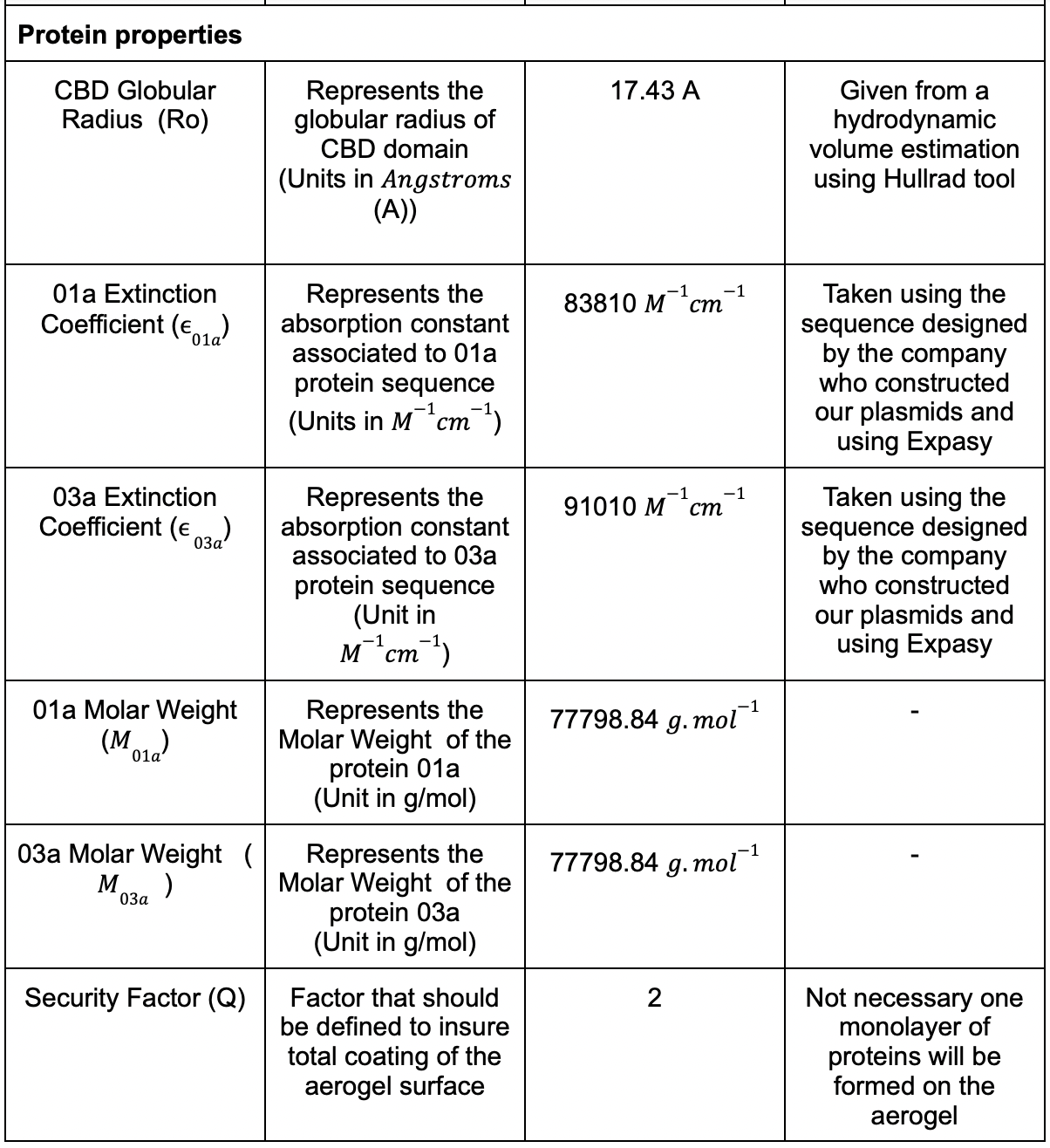
Table 1: Table for the parametric model .
In the proposed model we would like to coat geometrically our aerogel surface with different CBD globular entities. Since they are the most probable point of anchoring they will represent an entire protein in the counting of the amount of proteins. We supposed that the proteins would be placed in an adjacent manner.
First step was to consider the aerogel as a perfect cylinder that will be coated by the proteins. The surface of the cylinder is simplified to two circles and an elongated rectangle (fig2). We proceeded to a geometrical filling of the surface.

Figure 2 | Aerogel simplified representation as a perfect cylinder.
The attaching CBD region was taken from the above model 4JO5 to study its hydrodynamic characteristics using the Hullrad algorithm. This program was used to get specific useful information on the protein 4JO5:

Table 2 | Hullrad computation for CBD, AJO5
Using the values from (Table. 2), we populated the different surfaces of the aerogel with circular proteins using some simple geometry equations. The two values of AVSR and MD were studied in the example. Considering the protein radius and aerogel diameter, we came up with an iterative algorithm in three steps: Step 1: Populate the outer peripheral of the bigger cylinder:
We used the following angular equation to compute the angle of placement of the first small protein circle in the aerogel bigger circle:
alpha = 1 /π |arcsin(r(proteins)/(r(aerogel)- r(proteins)))*180| (1)
where r(proteins) and r(aerogel) represent respectively the radius of the proteins and the aerogel.
Step2: Compute the number of cylinders that can populate the peripheral of our the bigger circle:
nb(circles) =[360/(2*alpha)] (2)
where [x] is the greatest integer function s.
Step 3: Define a secondary circle that has the next small circles:
We iterated the new radius until we could not fit any small circles in the area (fig.12):
r(new)=r(previous)-( 2 *r(small)) (3)
where r(previous) is the large radius of the previous iteration.
Ending condition :
r(previous)< r(small)
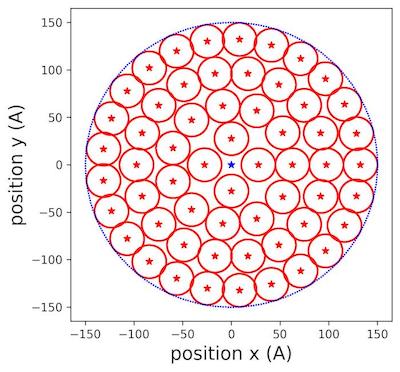
Figure 3 | Populating circles with tiny cylinders zoom at 150 Å aerogel surface .
In (fig.3), we saw clearly how the different circles were placed inside the allocated surface of aerogel. We extended the reasoning to filling the rectangular lateral surface.
For the lateral specific surface, the operation is described as following:
Step 1: Divide the width of the rectangle by the number of small protein circles that fits in the line :
nb(circles for width) = [W2 * r(proteins)] (4)
where W is the width of the rectangle, r(proteins) represents the radius of the proteins and [x] is the greatest integer function as described above.
Step 2: Multiply the above number by the number of lines that can fit in the height of the rectangle:
nb(circles for rectangle) = [h2* r(small)] * nb(circles for length) (5)
where h represents the height of the rectangle, [x] is the greatest integer function as described above.
By taking the parameters of the aerogel from (Table.1) and the parameters of the CBD from (Table.2) we got the following output in number of molecules and in number of molar:

Table 3: Results of the coating computations.
- The values found in (Table.3) were useful to define the protein solution volume that we soaked the hydrogels or the aerogels with.
Silk Specific Experiments Processes

Figure 4 | Protein Purification Protocol using Ni-NTA Beads
- For more information, check out our Protocols page on our wiki.

Figure 5 | Diagram of Procedure for Silk Biofilm Fabrication
- For more information, check out our Protocols page on our wiki.
Lab's Tips and Tricks
- Purification might prove difficult : elution with EDTA works better but might damage the protein more.
- The silk biofilm can be created even if the recombinant protein is in liquid form.
- When using formic acid, follow carefully the safety guidelines indicated on the Material Safety Datasheet.
Results
Bacterial Transformation
We followed the E.Coli competent cells quick protocol FB035 of Promega. We transformed BL21(DE3) cells with our plasmid containing mSA-N[AS]4C-CBD-10xHis. Cells were incubated on ice for about 10 min before heat shock. 400 µl of SOC medium was added after the heat shock. 100 µl of transformation reaction was plated on LB-Kana plates and the 300 µl left were put in 50 ml of LB-Kana in an Erlenmeyer to do liquid transformation cultures overnight at 37°C under shaking (200rpm).

Figure 6 | Bacterial transformation with the 01a construct in BL21(DE3) competent cells. (A) The plate transformed with 01a. (B) The plate transformed with a plasmid containing standard GFP (positive control). (C) The plate transformed with water (negative control).
After the overnight incubation, we observed the presence of colonies in the BL21(DE3) transformed cells plates of 01a and the positive control, no colonies were observed on the negative control, as expected.
. OD600 measurement: 2.914 was reached after the overnight incubation of the liquid transformation culture.
- Analysis
The 01a GeneScript plasmid transformation worked well for BL21(DE3) since we can observe colonies on the LB-Kanamycin plates and the OD600 of the transformation liquid culture is high, meaning that many bacteria were able to grow.
Protein Purification
The purification was performed following a Ni-NTA beads based protocol.

Figure 7 | Protein purification of silk fusion protein (01a)(A) SDS-PAGE gel stained with Coomassie Blue Protein Stain of all the fractions of silk fusion protein purification (B) Western Blot of the elution 1 (E1 fraction on SDS-PAGE gel) visualised with anti-His antibody. W1 = Wash 1 with 20 mM imidazole; W2 = Wash 2 with 50 mM imidazole; E1 = Elution 1 with 250 mM imidazole; E2 = Elution 2 with 500 mM imidazole; E4 = 2.5 M imidazole; E5 = 5 M imidazole.
- Analysis
Protein purification of the mSA-N[AS]4C-CBD-10xHis protein (01a) showed a 100kDa protein present in the first elution lane (E1) in both the SDS-PAGE (fig 7.A) and the Western Blot (fig 7.B). mSA-N[AS]4C-CBD-10xHis protein was expected at 78kDa size. Using the Nanodrop, we measured the concentration (0.72 mg/mL) and we obtained a final amount of 2.15 mg of the mSA-N[AS]4C-CBD-10xHis protein (01a). Since high imidazole concentration might denature the proteins, we had to either remove it or make imidazole inert. It was not possible to dialyse the proteins to remove imidazole, since the dialysis membrane was made of cellulose, and our fusion proteins would all bind to the membrane via their cellulose binding domain (CBD). We therefore decided to flash freeze the proteins and store them in the elution buffers so the imidazole will no longer affect them when frozen.
Cloning by digestion-ligation
The elution of the proteins of interest in such high amounts of imidazole certainly comes from the double His-tag that composes our proteins. To remove it from the received plasmids, we performed some cloning experiments to obtain better yields in the future purifications. In the case of the silk fusion protein, because of its repetitive modules the standard PCR-KLD cloning. To remove the added part, we therefore digested our plasmid (fig 8.A) with the NcoI enzyme since there are two NcoI restriction sites on both sides of the added site (fig 8.B) and re-ligated it back (fig 8.C).

Figure 8 | Cloning experiment results for the removal of the added site for 01a (silk). (A) Agarose gel electrophoresis of the digested product of 01a. (B) Plasmid map of 01a (silk) with the added part. (C) Plasmid map from the sequencing result of the obtained new 01a plasmid (silk).

Figure 9 | Expression of the silk fusion protein from the plasmid without the added site. (A) SDS-PAGE gel stained with Coomassie Blue Protein Stain of the lysate before and after IPTG induction. (B) SDS-PAGE gel stained with Coomassie Blue Protein Stain of the samples from protein purification of the new silk fusion protein induced in BL21(DE3). SN = Supernatant; FT = Flowthrough; W1 = Wash 1 with 20 mM imidazole; W2 = Wash 2 with 50 mM imidazole; E1 = Elution 1 with 250 mM imidazole; E2 = Elution 2 with 500 mM imidazole.
- Analysis
Since we obtained the wanted plasmid, we transformed new BL21(DE3) competent cells to start a new protein production. By doing a gel comparing the protein expression before and after IPTG induction (fig 9.A), we could see a band at approximately 100 kDa. Protein purification of the silk fusion protein (01a) also showed a 100kDa protein present in the two elution lanes (E1 and E2). Even though we expected the protein to be around 74kDa, this 100kDa lane should correspond to our new silk fusion protein, since we obtained similar ones in the previous purification. By measuring the concentration (0.37 mg/mL), we obtained a final amount of 11 mg of the silk fusion protein (01a)
Biofilm Fabrication

Figure 10 | Silk biofilms inside polystyrene petri dishes. (A) Silk fusion protein biofilm in polystyrene petri dish after 24 hours drying. (B) GFP fusion protein biofilm in polystyrene petri dish after 24 hours drying.
- Analysis
After the production of our proteins, we created a silk fusion protein biofilm to coat an aerogel to make it water-resistant. Inspired by the work of Felix Bauer, we produced two biofilms: one with the recombinant silk protein (fig 10.A) and one with the recombinant GFP protein, as a control (fig 10.B). In figure 8, we can see that we dried the biofilms too much due to the presence of cracks. Even if it was not a problem for hydrophobicity testing, it became one to remove the biofilm from the dish. Indeed, it was impossible for us to take the biofilm off, even by cutting the edges of the petri dish. This meant that we couldn’t coat aerogels with those biofilms. However, we saw that the GFP biofilm (fig 10.B) made some crystals and cubic structures so that the intended biofilm was less homogeneous than the silk one (fig 10.A). Overall, we managed to generate a biofilm with our recombinant silk fusion protein (mSA-silk-CBD), but not with the GFP fusion protein (mSA-GFP-CBD).
Hydrophobicity Test
With these biofilms, we did a wettability test to compare their hydrophobicity. To do so, we added droplets of water on top of those biofilms and did several measurements.

Figure 11 | Silk biofilms with water droplets for hydrophobicity tests. (A) Silk fusion protein biofilm (01a) with a drop of water (B) GFP fusion protein biofilm (01a) with a drop of water. (C) Measurement of the angle made by the drop on the silk biofilm. (D) Measurement of the angle made by the drop on the GFP biofilm. (E) Dried drops of water on the silk biofilm (F) Dried drops of water on the GFP biofilm.
- Analysis
First, we measured the angle made by the droplet (fig 11.A and B) to assess surface tension and the surface wettability. Then we measured the time it took for the water to be absorbed or evaporated, computed the mean values for each biofilm and compared them to conclude about hydrophobicity. For the first measures, we obtained an angle of 55.2° for the silk (fig 11.C) and 27.3° for the GFP biofilm (fig 11.D). This meant that the silk fusion protein biofilm (01a) had a higher contact angle than the GFP biofilm. Moreover, by measuring the time it takes for three water droplets to be absorbed, we obtained a mean of 466,33 s for the silk biofilm (fig 11.E), and 28,33 s for the GFP biofilm (fig 11.F). With these values, we concluded that it takes more time for a drop of water to be evaporated or absorbed from the silk biofilm. These two results suggested that our biofilm helps protect the aerogel from water droplets.
Coating an aerogel with mSA-N[AS]4C-CBD-10xHis protein
We showed that coating the aerogel with the silk fusion protein makes it more water resistant than without the coating. We first coated hydrogels with proteins (either the silk fusion protein (01a), the GFP fusion protein (03a), the GFP control protein or water) (fig 12.A) and sent them to freeze dry (fig 12.B). After this step, we saw that the coated aerogels had a more homogeneous shape, with less cracks and a sort of protective film on top of it compared to the uncoated ones (fig 13.A).

Figure 12 | Freeze drying of coated hydrogels. (A) Hydrogels before freeze drying. (B) Aerogel after 23 hours of freeze drying.

Figure 13 | Protective coating given by the silk fusion protein. (A) Comparison between uncoated and coated with recombinant silk (01a) aerogels. (B) Boxplots of the delay time a drop of water gets absorbed inside an aerogel coated with the different proteins. Top of the boxes = mean of delay time per group; Black dots = individual delay time measurements; Red vertical line = standard error of the mean. Data are mean ± s.d., n = 3 measurements per group; ***P < 0.001, **P < 0.01; one way ANOVA test was performed followed by a Tukey HSD for performing multiple pairwise-comparisons between the means of groups.
- Analysis
Using the Keyence microscope, we filmed the application of 4 to 5 drops of water on the aerogels and quantified the water absorption time. For each aerogel we computed the mean delay time and used this data to perform a one-way ANOVA test to see if there is a significant difference between the means between the groups. We obtained a final p-value of 0.000576. As the p-value was less than the significance level 0.05, we concluded that there are significant differences between the groups. However, this result did not indicate which pairs of groups are different. As the ANOVA test was significant, we computed a Tukey HSD for performing multiple pairwise-comparisons between the means of groups. For each comparison with the silk fusion protein (01a), we obtained a p-value way smaller than 0.05, which means that there was a significant difference between the silk fusion protein and the other groups, while there was no significant difference among the other groups (dH20, GFP and GFP fusion protein 03a). Therefore we concluded that aerogels absorb water more slowly when they are coated with the silk fusion protein. Indeed, in the box plot we plotted to present the different populations (fig 13.B), it is clear that in the case of the silk fusion protein (01a), the delay times were higher. The water drop took longer to be fully absorbed by the aerogel coated with the recombinant silk, suggesting that the recombinant silk adds hydrophobicity to the aerogel. Overall, these results proved that the CBD domain we added to our silk construct allows binding to the aerogel and that this protein provides a protective coating for the aerogel.
References
- Bauer, F. & Scheibel, T. (2012, 16 mai). Artificial Egg Stalks Made of a Recombinantly Produced Lacewing Silk Protein. Angewandte Chemie International Edition, 51(26), 6521‑6524. https://doi.org/10.1002/anie.201200591
- Bauer, F. (2013) Development of an artificial silk protein on the basis of a lacewing egg stalk protein. https://epub.uni-bayreuth.de/72/1/Diss.pdf
- Lim, K. H., Huang, H., Pralle, A. & Park, S. (2012, août 8). Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnology and Bioengineering, 110(1), 57‑67. https://doi.org/10.1002/bit.24605
