Difference between revisions of "Part:BBa K112000"
(→Characterized by CAFA_China 2022) |
|||
| (4 intermediate revisions by one other user not shown) | |||
| Line 86: | Line 86: | ||
[https://2020.igem.org/Team:SZ-SHD/Experiment SZ-SHD/wiki/experiment] | [https://2020.igem.org/Team:SZ-SHD/Experiment SZ-SHD/wiki/experiment] | ||
| − | + | ==Team 2022 SZ-SHD== | |
| − | == | + | ===Characterization of arabinose induced self- lysis system=== |
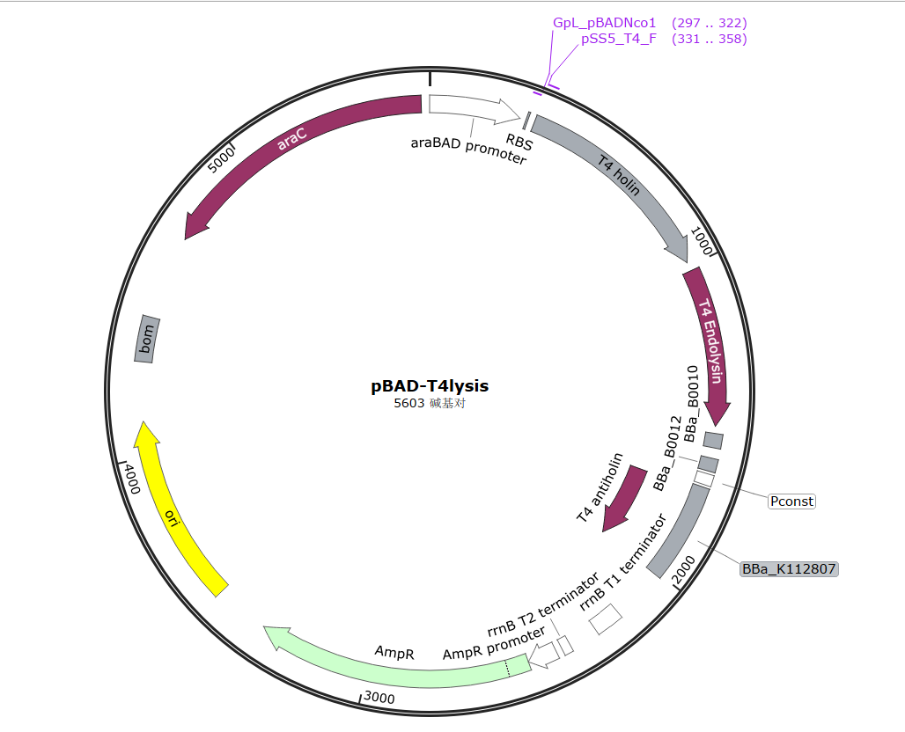
| − | + | To co-transform the self-lysis system with our OMV-producing vector(pSB1C3 backbone), the antibiotic resistance tag has to be changed(two tags need to be different), therefore, we use pBAD-HisA plasmid as our backbone. | |
| − | + | [[File:T--SZ-SHD--000.png|600px|thumb|center|Fig3 Construction of self-lysis vector]] | |
| + | After checking more instructions about araBAD promoter, we found that the arabinose concentration used in 2020(1mM/L≈0.15g/L) is far lower than the manual suggest(2g/L)(M.R. Green, Molrcular cloning a laboratory manual (fourth edition)), we improved our protocol and accelerate the lysis process from 20 hour to 4 hour. | ||
| + | [[File:T--SZ-SHD--001.png|600px|thumb|center|Fig4. OD600 change after 1mmol/L final concentration of arabinose added]] | ||
| + | [[File:T--SZ-SHD--002.png|600px|thumb|center|Fig5. OD600 change after 2g/L final concentration of arabinose added]] | ||
| + | (more information: https://2022.igem.wiki/sz-shd/contribution) | ||
Latest revision as of 12:05, 12 October 2022
T4 holin, complete CDS, berkeley standard
T4 holin gene, with start and stop codons.
This part is in BBb Format. It is flanked by BamHI and BglII sites instead of XbaI and SpeI. More information about the BBb Format is available at:
[http://openwetware.org/wiki/Template:AndersonLab:BBb_Standard BBb Standard Description Page]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team NYMU-Taipei 2017: Successfully construct T4 holin into suicide mechanism
Improvement
We successfully combine T4 holin with a lactose-induced promoter (BBa_R0010), a ribosome binding site (BBa_B0034) and a double terminator (BBa_B0010 and BBa_B0012). Therefore, holin can be induced by lactose and regulate the suicide mechanism. Moreover, we construct T4 holin and T4 endolysin together as a functional suicide mechanism. Besides, in NYMU-Taipei 2017 team’s project, we also put the suicide mechanism into practice by constructing them with NrtA.
Result
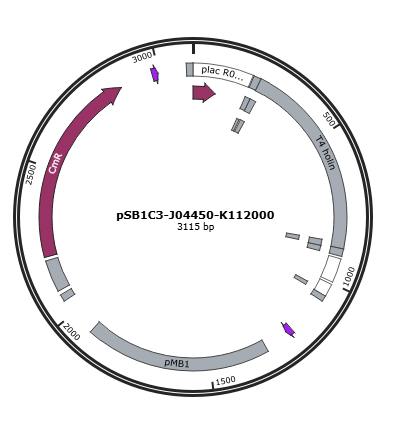
Figure 1 is the gene map of T4 holin construct, pSB1C3-J04450-K112000.
Figure 1
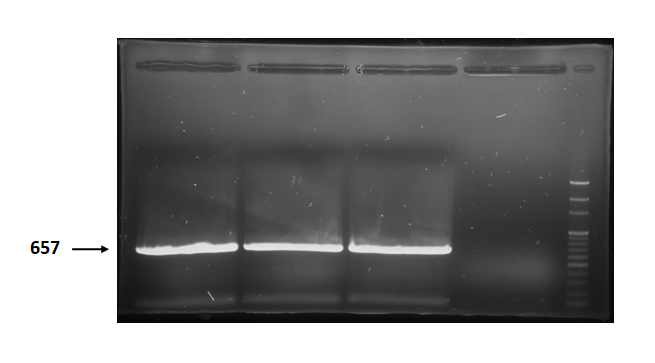
Figure 2 is electrophoresis result of holin (from BBa_K112000) PCR product.
The marker is 100bp. The length is 657bp as expected.
Figure 2
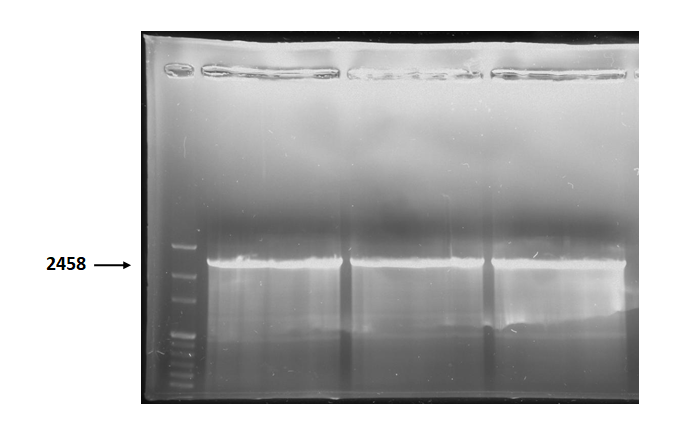
Figure 3 is electrophoresis result of holin backbone (from BBa_J04450) PCR product.
The marker is 1kb. The length is 2458bp as expected.
Figure 3
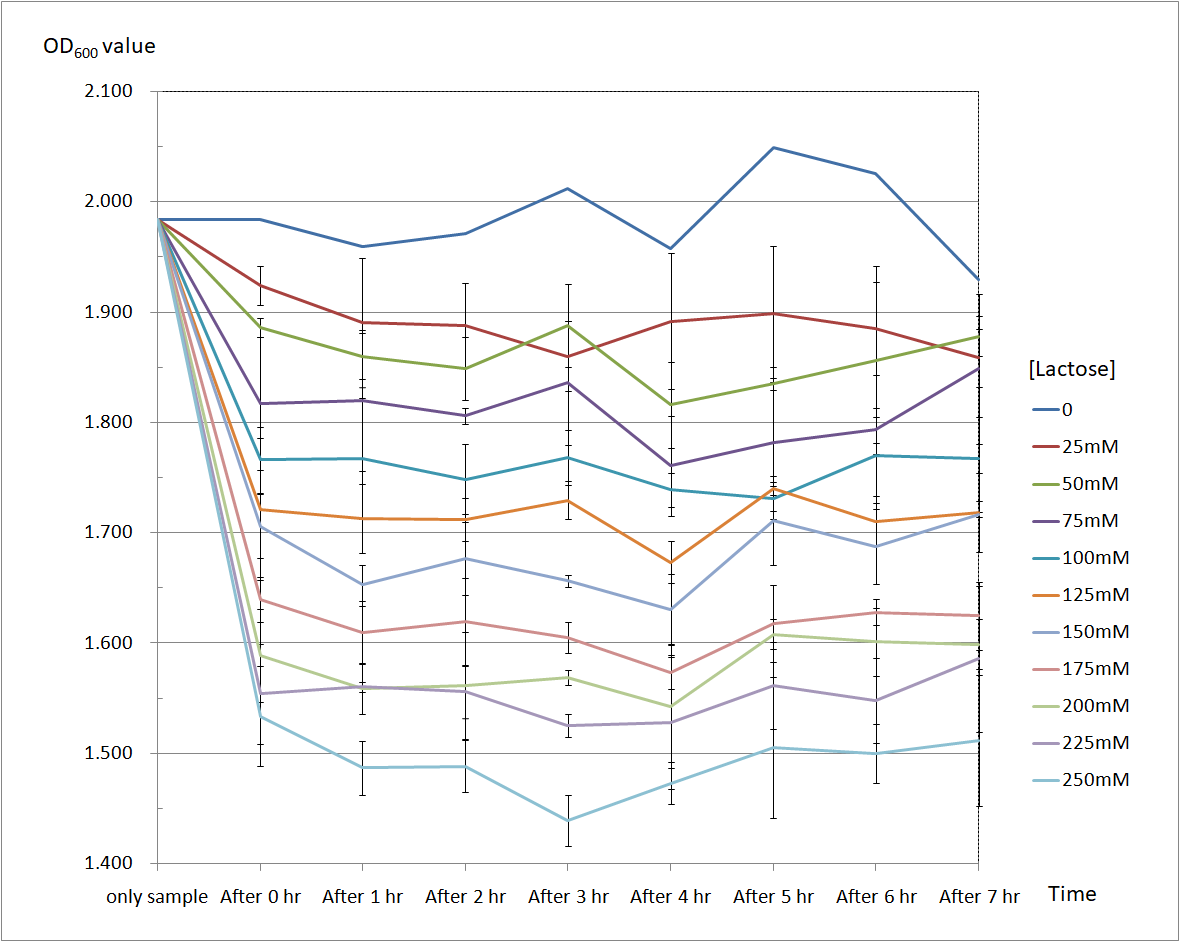
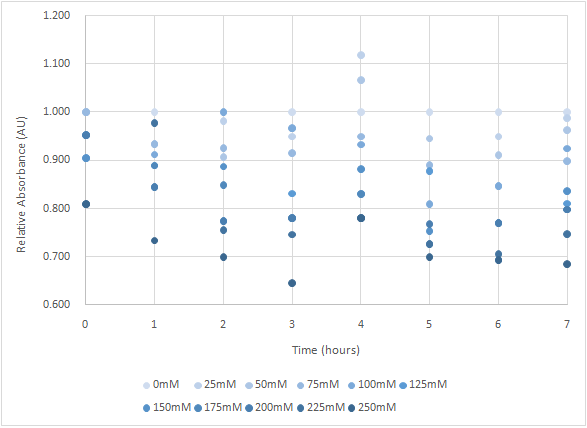
Figure 4 and Figure 5 are the results of suicide mechanism functional test. Both figure show that our suicide mechanism can be induced by adding lactose, and the effectiveness of suicide mechanism goes better as the concentration of lactose goes higher.
Figure 4 is the bacterium with holin-endolysin construct.
As figure 3 shows, the suicide mechanism is induced immediately when lactose is added into the samples. Besides, we can see that the lactose concentration and the OD value of the bacterium with holin-endolysin construct are positively correlated, which means our suicide mechanism does work.
Figure 4
Figure 5 is the bacterium with holin-endolysin-NrtA construct.
As figure 5 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance.
Figure 5
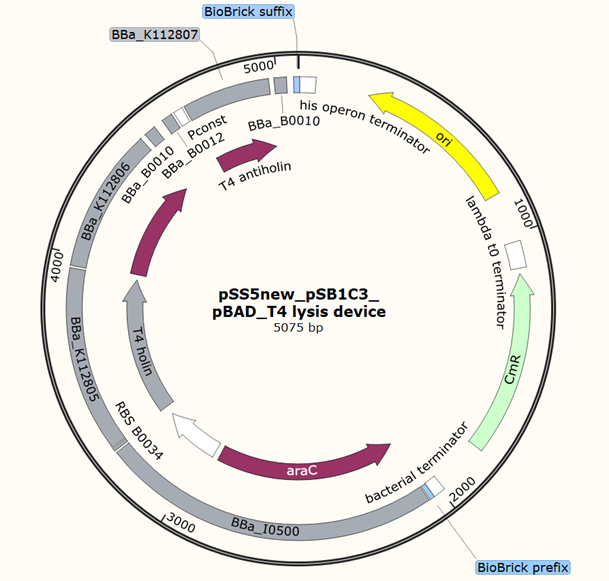
Team 2020 SZ-SHD
Application of pBAD, L-arabinose-inducible promoter, to regulate T4 holin and T4 endolysin production as a cell suicide switch. Which has been applied to control the release of cytoplasmic proteins.
Plasmid circuit map:
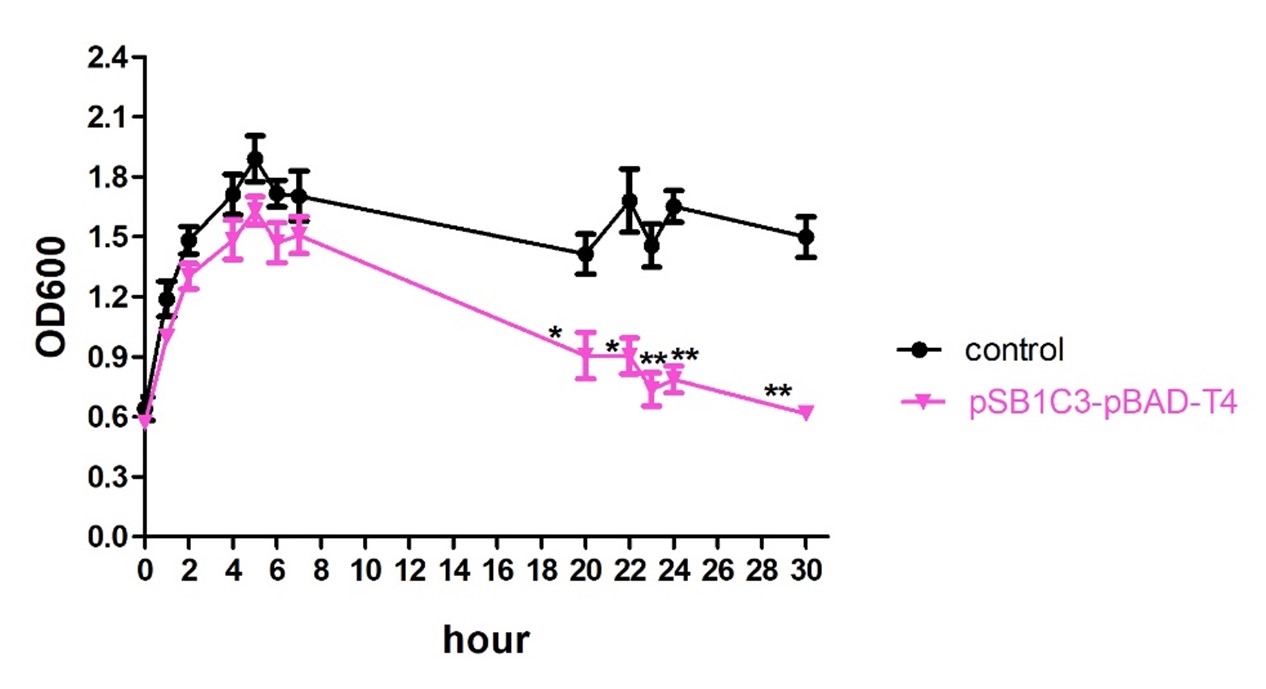
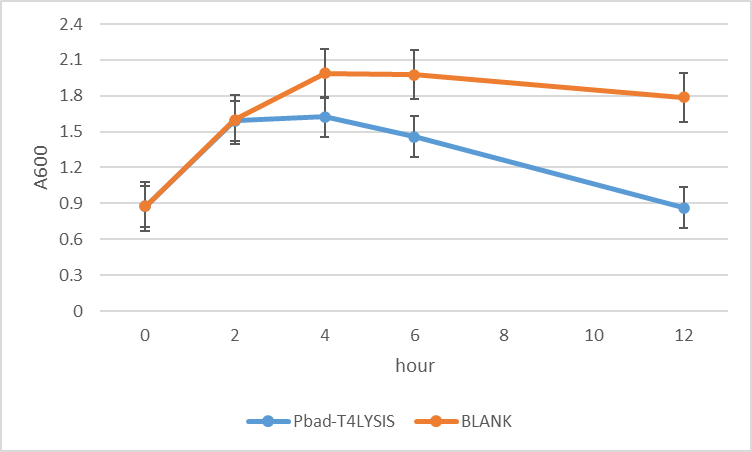
The experiment of the suicide efficiency was investigated using E. coli (strain DH10B), a gradual decline in OD600 value of recombined DH10B (pSB1C3-pBAD-T4) in response to 1mM arabinose could be observed comparing to non-transformed ones (control).
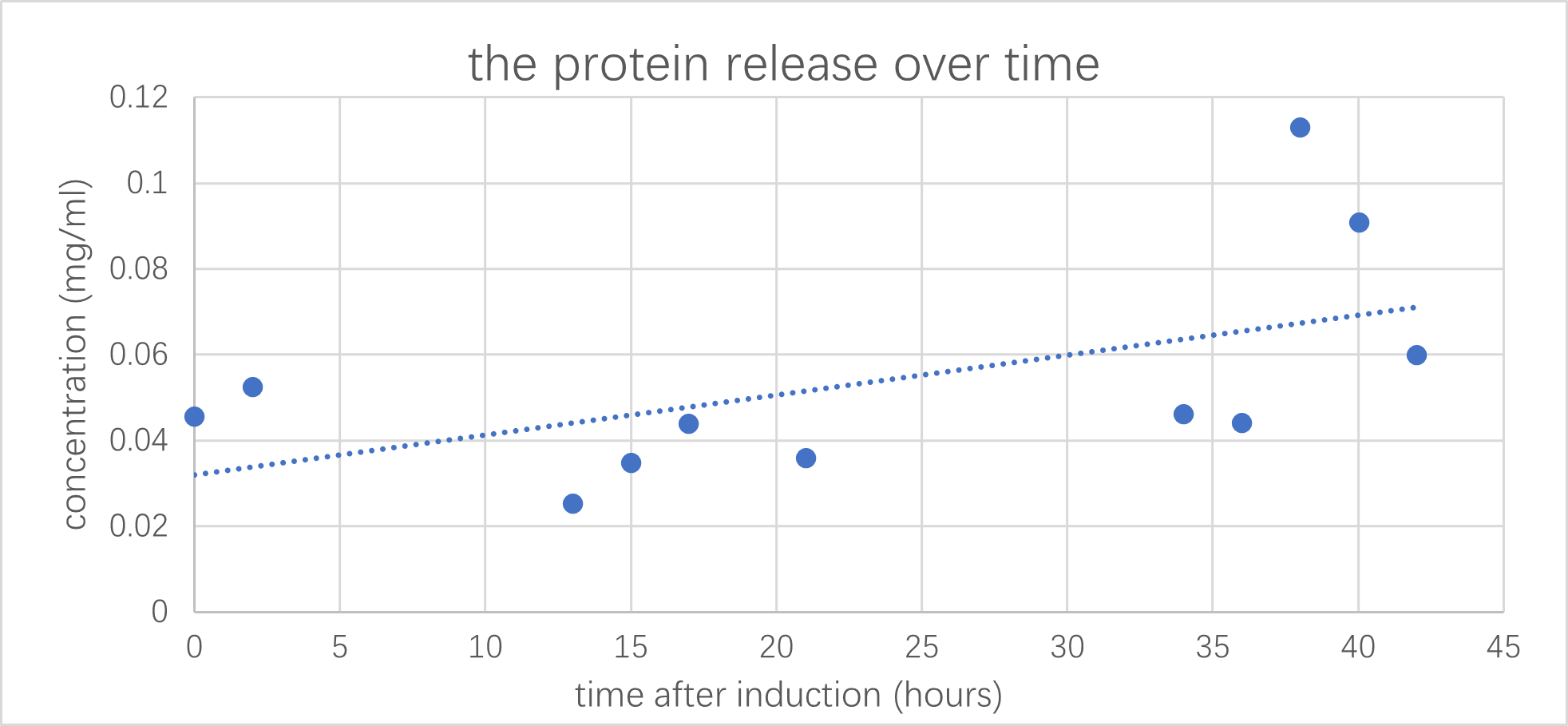
We have co-transformed this plasmid with a protein expressing circuit, which we have observed a increase in protein concertation after 1mM of arabinose has been added.
more about how we use this part:
SZ-SHD/wiki/part
SZ-SHD/wiki/experiment
Team 2022 SZ-SHD
Characterization of arabinose induced self- lysis system
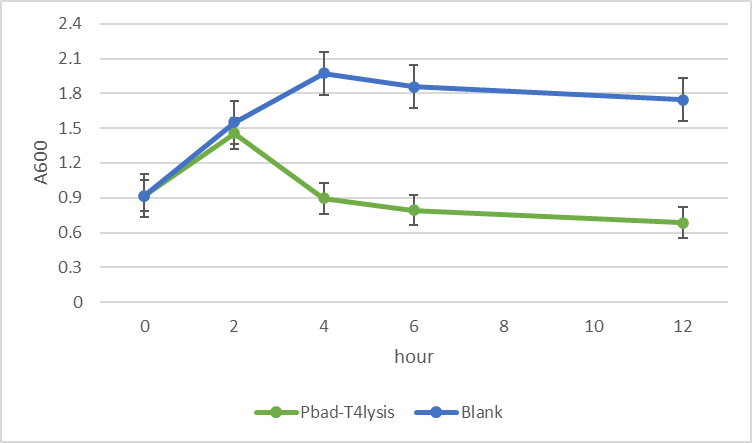
To co-transform the self-lysis system with our OMV-producing vector(pSB1C3 backbone), the antibiotic resistance tag has to be changed(two tags need to be different), therefore, we use pBAD-HisA plasmid as our backbone.
After checking more instructions about araBAD promoter, we found that the arabinose concentration used in 2020(1mM/L≈0.15g/L) is far lower than the manual suggest(2g/L)(M.R. Green, Molrcular cloning a laboratory manual (fourth edition)), we improved our protocol and accelerate the lysis process from 20 hour to 4 hour.
(more information: https://2022.igem.wiki/sz-shd/contribution)