Difference between revisions of "Part:BBa K4428001"
Dhwaniteck (Talk | contribs) |
Dhwaniteck (Talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K4428001 SequenceAndFeatures</partinfo> | <partinfo>BBa_K4428001 SequenceAndFeatures</partinfo> | ||
| + | |||
==Background== | ==Background== | ||
For the biosorption of lead ions, PbrR has been effectively engineered onto the surface of microorganisms. However, this molecule's lack of specificity makes it challenging to recover a single heavy metal ion selectively. The PbrR coming from the MerR family was characterized structurally by two distinct domains: an N-terminal DNA-binding domain (DBD) and a C-terminal metal binding domain (MBD) containing a central linker. Studies claimed that the MerR member was made up of the C-terminal conserved MBD and the N-terminal distinct DBD (Chang et al. 2015). It is known that the smaller the molecule the better the cell surface expression. Also, studies showed that both the domain’s functions are independent of each other (Song et al. 2004; Tao et al. 2013). This led to the expression of just the metal-binding-domain on the cell surface to increase the surface hence increasing the adsorption capacity.<br><br> | For the biosorption of lead ions, PbrR has been effectively engineered onto the surface of microorganisms. However, this molecule's lack of specificity makes it challenging to recover a single heavy metal ion selectively. The PbrR coming from the MerR family was characterized structurally by two distinct domains: an N-terminal DNA-binding domain (DBD) and a C-terminal metal binding domain (MBD) containing a central linker. Studies claimed that the MerR member was made up of the C-terminal conserved MBD and the N-terminal distinct DBD (Chang et al. 2015). It is known that the smaller the molecule the better the cell surface expression. Also, studies showed that both the domain’s functions are independent of each other (Song et al. 2004; Tao et al. 2013). This led to the expression of just the metal-binding-domain on the cell surface to increase the surface hence increasing the adsorption capacity.<br><br> | ||
| − | Importantly, the | + | Importantly, the Pb<sup>2+</sup> adsorption capacity of PbrR MBD-displayed cells was about 1.92-fold higher than that of the full-length PbrR-displayed cells. Also, the PbrR MBD shows highly selective adsorption towards the lead similar to the full-length PbrR (Hui et al. 2018). |
| + | |||
| + | [[File:IITD_PbrR_MBD_Structure.png|800px|thumb|centre|A model of PbrR protomer generated using Swiss-Model workspace (https://swissmodel.expasy.org/workspace/). The ribbon diagram shows the Pb<sup>2+</sup> binding domain in color and the other parts in gray. Residues Cys79, Cys114, and Cys123 involved in metal binding are presented as a space-filling model. d Ribbon representation of PbBD generated by truncating 49 N-terminal amino acids and 15 C-terminal amino acids of PbrR]] | ||
==Part Uses== | ==Part Uses== | ||
In contrast to the conventional physical and chemical methods for environmental remediation, microbial cell-surface display has evolved as a unique strategy for the bioremediation of heavy metas. An innovative method for the selective bioremediation of harmful heavy metals is the engineering of the particular metal binding domain or motif derived from the MerR family regulators on the microbial cell surface. <br><br> | In contrast to the conventional physical and chemical methods for environmental remediation, microbial cell-surface display has evolved as a unique strategy for the bioremediation of heavy metas. An innovative method for the selective bioremediation of harmful heavy metals is the engineering of the particular metal binding domain or motif derived from the MerR family regulators on the microbial cell surface. <br><br> | ||
| − | The PbrR MBD domain due to its smaller size can lead to increased cell surface display causing better adsorption of the metal ion. Using the MBD instead of the full length PbrR for expression on cell surface will prove better for bioremediation of the heavy metal | + | The PbrR MBD domain due to its smaller size can lead to increased cell surface display causing better adsorption of the metal ion. Using the MBD instead of the full length PbrR for expression on cell surface will prove better for bioremediation of the heavy metal Pb<sup>2+</sup> ions. |
| + | |||
| + | [[File:IITD_PbrR_MBD_Display.png|600px|thumb|centre|Surface display of PbrR and PbBD fusions to LOA in E. coli BL21(DE3). a Bright field and corresponding fluorescent micrographs of surface-engineered cells. Immunofluorescence labelling of E. coli cells using anti-His tag antibody (primary antibody) and FITC-labeled donkey anti-mouse antibody (secondary antibody). b Immunofluorescence intensity of full-length PbrR or PbBD-displayed E. coli cells. Data are the mean of three independent measurements]] | ||
| + | |||
| + | ===Improvement over PbrR for Lead Recovery=== | ||
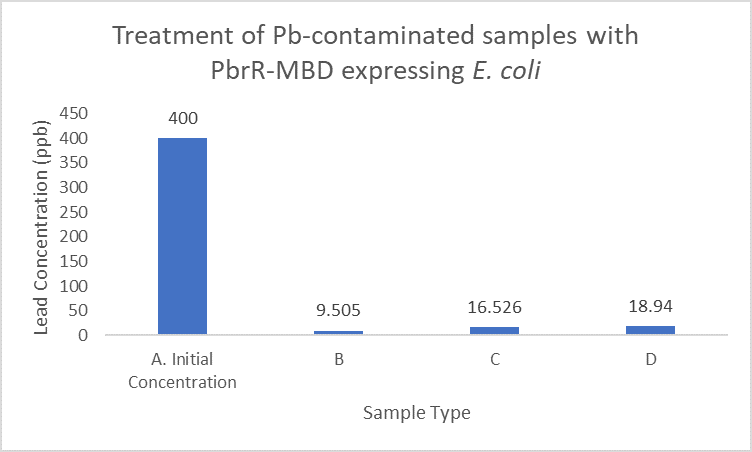
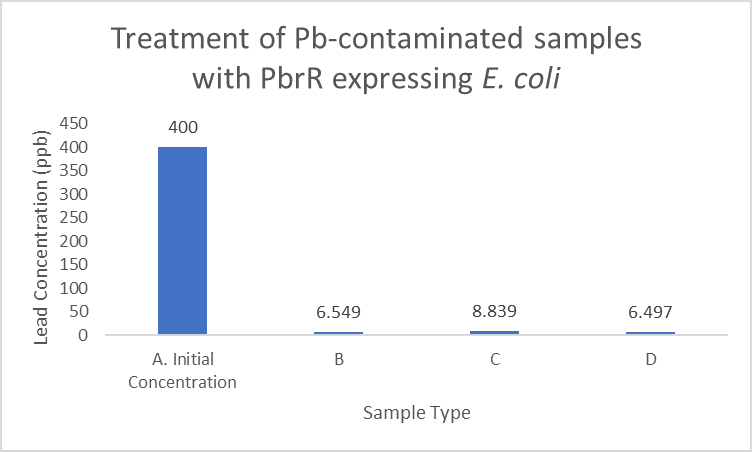
| + | When subject to the same 400 ppb concentration of lead, PbrR MBD is able to remove as much amount of lead from the system as PbrR. This clearly suggests that even this smaller protein is sufficient for lead adsorption at 0.4 mg/l concentration of lead which is the concentration of lead in industrial effluents. ICP-MS results suggest that this is nowhere near the maximum capacity of adsorption of the proteins which would actually be much more than what is currently tested. However, this in itself suggests that even this smaller protein can work as well as PbrR in these concentrations and thus, reduces the metabolic burden of the cell. | ||
| + | |||
| + | [[File:IITD_PbrR_MBD_Treatment.png|600px|thumb|centre|Sample A represents the initial concentration of the artificially contaminated water. Samples B, C and D are the contaminated samples treated with PbrR-MBD expressing cells for 6 hours. The bacteria were induced for different durations before the treatment. Sample D is treated with uninduced cells: A. Original concentration of lead-contaminated sample, B. Contaminated sample treated with PbrR-MBD expressing bacteria which were induced for 6 hours, C. Contaminated sample treated with PbrR-MBD expressing bacteria which were induced for 12 hours (overnight induction), D. Contaminated sample treated with uninduced PbrR-MBD expressing bacteria]] | ||
| + | |||
| + | [[File:IITD_PbrR_Treatment.png|600px|thumb|centre|Sample A represents the initial concentration of the artificially contaminated water. Samples B, C and D are the contaminated samples treated with PbrR-expressing cells for 6 hours. The bacteria were induced for different durations before the treatment. Sample D is treated with uninduced cells: A. Original concentration of lead-contaminated sample, B. Contaminated sample treated with PbrR expressing bacteria which were induced for 6 hours, C. Contaminated sample treated with PbrR expressing bacteria which were induced for 12 hours (overnight induction), D. Contaminated sample treated with uninduced PbrR expressing bacteria]] | ||
| + | |||
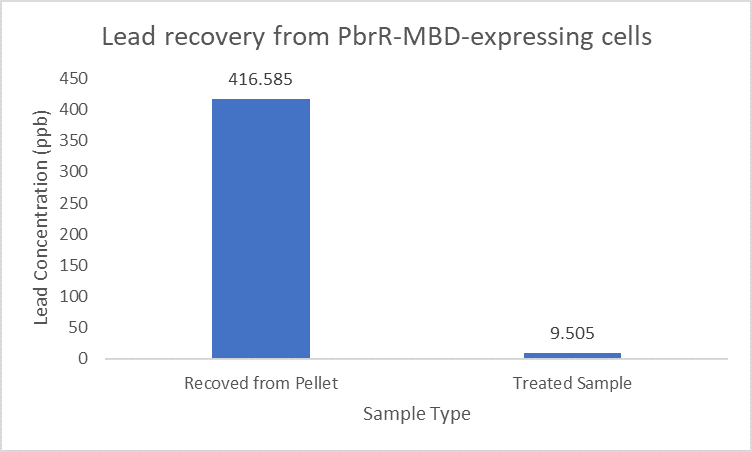
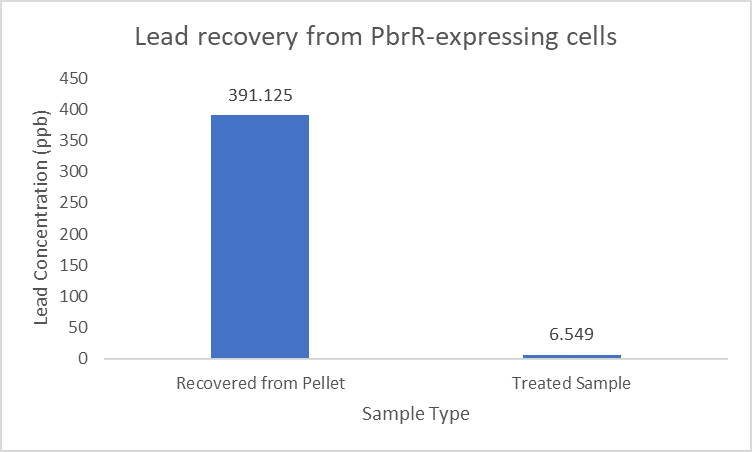
| + | Further, it observe that a greater amount of lead is successfully desorbed from the cells expressing PbrR-MBD on their surface than PbrR. This suggests that PbrR-MBD may have an even greater ability to recover lead than PbrR. This observation holds great significance when thinking about the recycling of lead instead of just its removal. | ||
| + | |||
| + | [[File:IITD_PbrR_MBD_Recovery.png|600px|thumb|centre|]] | ||
| + | [[File:IITD_PbrR_Recovery.png|600px|thumb|centre|]] | ||
Latest revision as of 15:18, 12 October 2022
PbrR MBD - Lead-binding domain of PbrR
PbrR was modified by truncating the N-terminal DNA Binding Domain and C-terminal redundant amino acid residues to obtain lead domain maintaining its Pb2+ binding property. The MBD (metal binding domain) being smaller in size leads to increased display and expression than the full-length PbrR-displayed cells.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 229
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 229
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 229
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 229
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 20
Background
For the biosorption of lead ions, PbrR has been effectively engineered onto the surface of microorganisms. However, this molecule's lack of specificity makes it challenging to recover a single heavy metal ion selectively. The PbrR coming from the MerR family was characterized structurally by two distinct domains: an N-terminal DNA-binding domain (DBD) and a C-terminal metal binding domain (MBD) containing a central linker. Studies claimed that the MerR member was made up of the C-terminal conserved MBD and the N-terminal distinct DBD (Chang et al. 2015). It is known that the smaller the molecule the better the cell surface expression. Also, studies showed that both the domain’s functions are independent of each other (Song et al. 2004; Tao et al. 2013). This led to the expression of just the metal-binding-domain on the cell surface to increase the surface hence increasing the adsorption capacity.
Importantly, the Pb2+ adsorption capacity of PbrR MBD-displayed cells was about 1.92-fold higher than that of the full-length PbrR-displayed cells. Also, the PbrR MBD shows highly selective adsorption towards the lead similar to the full-length PbrR (Hui et al. 2018).

Part Uses
In contrast to the conventional physical and chemical methods for environmental remediation, microbial cell-surface display has evolved as a unique strategy for the bioremediation of heavy metas. An innovative method for the selective bioremediation of harmful heavy metals is the engineering of the particular metal binding domain or motif derived from the MerR family regulators on the microbial cell surface.
The PbrR MBD domain due to its smaller size can lead to increased cell surface display causing better adsorption of the metal ion. Using the MBD instead of the full length PbrR for expression on cell surface will prove better for bioremediation of the heavy metal Pb2+ ions.

Improvement over PbrR for Lead Recovery
When subject to the same 400 ppb concentration of lead, PbrR MBD is able to remove as much amount of lead from the system as PbrR. This clearly suggests that even this smaller protein is sufficient for lead adsorption at 0.4 mg/l concentration of lead which is the concentration of lead in industrial effluents. ICP-MS results suggest that this is nowhere near the maximum capacity of adsorption of the proteins which would actually be much more than what is currently tested. However, this in itself suggests that even this smaller protein can work as well as PbrR in these concentrations and thus, reduces the metabolic burden of the cell.
Further, it observe that a greater amount of lead is successfully desorbed from the cells expressing PbrR-MBD on their surface than PbrR. This suggests that PbrR-MBD may have an even greater ability to recover lead than PbrR. This observation holds great significance when thinking about the recycling of lead instead of just its removal.