Difference between revisions of "Part:BBa K4140015"
Ahmed Mattar (Talk | contribs) |
(→Usage) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
==Part Description== | ==Part Description== | ||
| − | A common structural motif identified in functioning RNA species is the kink-turn (k-turn). It typically consists of a tandem trans sugar edge-Hoogsteen G followed by a three-nucleotide bulge: pair A bases. The minor grooves are juxtaposed and the axis of duplex RNA is given a strong bend. The conserved adenine nucleobases of the G:A | + | A common structural motif identified in functioning RNA species is the kink-turn (k-turn). It typically consists of a tandem trans sugar edge-Hoogsteen G followed by a three-nucleotide bulge: pair A bases. The minor grooves are juxtaposed and the axis of duplex RNA is given a strong bend. The conserved adenine nucleobases of the G:A base pairs accept the cross-strand H-bonds that form at the interface. The k-turns are split into two conformational classes, N3 and N1, by alternative acceptors for one of these. The confirmation that a specific k-turn adopts is determined by the base pair (3b:3n) that follows the G:A pairings. K-turns typically bind proteins and mediate tertiary contacts in folded RNA species. Members of the L7Ae family of proteins are frequently found to bind k-turns |
| + | |||
==Usage== | ==Usage== | ||
| − | + | We employed K-turns' capacity to mediate tertiary contacts in folded RNA species and bind proteins. As a result, we employ it to regulate the expression of our CRISPR regulatory system since, when combined with L7Ae, it suppresses the translation of the protein portion (Cas12g), Whenever there is a high quantity of phenylalanine as shown in figure 1. | |
| − | + | ||
[[Image:reg.png|thumb|right|Figure(1) Shows an SBOL demonstrating the usage of k-turns in our whole cel-based biosensor ]] | [[Image:reg.png|thumb|right|Figure(1) Shows an SBOL demonstrating the usage of k-turns in our whole cel-based biosensor ]] | ||
| + | <br><br><br><br><br><br><br><br><br><br> | ||
==Literature Characterization== | ==Literature Characterization== | ||
| Line 17: | Line 18: | ||
[[File:Kt.png|thumb|Right|Figure 1. Analysis of ion-induced folding of Kt-23 and variants studied by FRET.]] | [[File:Kt.png|thumb|Right|Figure 1. Analysis of ion-induced folding of Kt-23 and variants studied by FRET.]] | ||
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| − | + | ==Characterization by mathematical modeling== | |
| + | This model is to simulate the kinetics of the riboswitch (L7Ae with kink turns) that is used in our circuit. The designed circuit is to detect if the increasing substance is either phenylalanine or tyrosine via TyrR. So if phenylalanine level is elevated, L7Ae is formed as it is downstream TyrR that forms a complex via binding with its kink-turn on another circuit; that complex inhibits expression of cas12g, so the circuit will be able to express lacZ alpha (beta-galactosidase) in diagnostic circuit or PAH in the therapeutic circuit. If tyrosine level is elevated with a decreased level of phenylalanine, it activates tyrR inhibitory promoter so no L7Ae would be expressed resulting in cas12g expression to control the circuit as shown figure (4) and graph (1). | ||
| + | [[File:rib11.png|Right|]] | ||
| + | <br><br> | ||
| + | Figure (4) illustrates the kinetics of all reactions in riboswitch model | ||
| + | [[File:rib22.png|Right|]] | ||
| + | <br><br> | ||
| + | Graph (1) illustrates riboswitch kinetics in which Q represents the condition where L7Ae is expressed and bound to its kink-turns ,therefore inhibiting the expression of cas12g. However, M represents no expression of L7Ae in which cas12g would be expressed to control the circuit if the phenylalanine is absent. | ||
| + | ==Experimental Characterization== | ||
| + | [[File:capture7.png|right|]] | ||
| + | <br><br><br><br><br> | ||
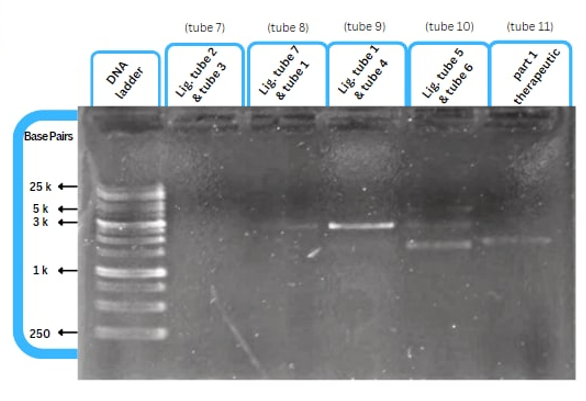
| + | This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 4. The running part (ordered from IDT) included Human u6 Promoter - -Kinkturn - CMV Promoter - Cas12g. | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
==References== | ==References== | ||
Latest revision as of 19:06, 11 October 2022
Kink turn
Part Description
A common structural motif identified in functioning RNA species is the kink-turn (k-turn). It typically consists of a tandem trans sugar edge-Hoogsteen G followed by a three-nucleotide bulge: pair A bases. The minor grooves are juxtaposed and the axis of duplex RNA is given a strong bend. The conserved adenine nucleobases of the G:A base pairs accept the cross-strand H-bonds that form at the interface. The k-turns are split into two conformational classes, N3 and N1, by alternative acceptors for one of these. The confirmation that a specific k-turn adopts is determined by the base pair (3b:3n) that follows the G:A pairings. K-turns typically bind proteins and mediate tertiary contacts in folded RNA species. Members of the L7Ae family of proteins are frequently found to bind k-turns
Usage
We employed K-turns' capacity to mediate tertiary contacts in folded RNA species and bind proteins. As a result, we employ it to regulate the expression of our CRISPR regulatory system since, when combined with L7Ae, it suppresses the translation of the protein portion (Cas12g), Whenever there is a high quantity of phenylalanine as shown in figure 1.
Literature Characterization
In this study, Using comparative gel electrophoresis, When Kt-7 and Kt-23 undergo electrophoresis side by side in the presence of 2 mM Mg2+ ions, their mobility patterns are very comparable, indicating that they fold to the same extent under these circumstances.
Characterization by mathematical modeling
This model is to simulate the kinetics of the riboswitch (L7Ae with kink turns) that is used in our circuit. The designed circuit is to detect if the increasing substance is either phenylalanine or tyrosine via TyrR. So if phenylalanine level is elevated, L7Ae is formed as it is downstream TyrR that forms a complex via binding with its kink-turn on another circuit; that complex inhibits expression of cas12g, so the circuit will be able to express lacZ alpha (beta-galactosidase) in diagnostic circuit or PAH in the therapeutic circuit. If tyrosine level is elevated with a decreased level of phenylalanine, it activates tyrR inhibitory promoter so no L7Ae would be expressed resulting in cas12g expression to control the circuit as shown figure (4) and graph (1).

Figure (4) illustrates the kinetics of all reactions in riboswitch model

Graph (1) illustrates riboswitch kinetics in which Q represents the condition where L7Ae is expressed and bound to its kink-turns ,therefore inhibiting the expression of cas12g. However, M represents no expression of L7Ae in which cas12g would be expressed to control the circuit if the phenylalanine is absent.
Experimental Characterization
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 4. The running part (ordered from IDT) included Human u6 Promoter - -Kinkturn - CMV Promoter - Cas12g.
References
1. Schroeder, K. T., & Lilley, D. M. (2009). Ion-induced folding of a kink turn that departs from the conventional sequence. Nucleic acids research, 37(21), 7281-7289. Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]