Difference between revisions of "Part:BBa K4192120"
| (9 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K4192120 short</partinfo> | <partinfo>BBa_K4192120 short</partinfo> | ||
| − | The expression products of obcA and obcB are enzymes needed to produce oxalate from citric acid and acetyl-CoA. And FpOAR protein | + | The expression products of <i>obcA</i> and <i>obcB</i> are enzymes needed to produce oxalate from citric acid and acetyl-CoA. And FpOAR protein can efflux oxalate. The composite part of these three genes can efflux oxalic acid out of the thallus. |
Because it only have one promoter at the beginning of the whole genetic circuit, the efflux rate of the generated oxalic acid is relatively low, which may also lead to the death of the bacteria more easily due to excessive accumulation of oxalic acid in the body. | Because it only have one promoter at the beginning of the whole genetic circuit, the efflux rate of the generated oxalic acid is relatively low, which may also lead to the death of the bacteria more easily due to excessive accumulation of oxalic acid in the body. | ||
| − | We have already tested it in E.coli and Pseudomonas fluorescens. | + | We have already tested it in <i>E.coli</i> and <i>Pseudomonas fluorescens</i>. |
| + | |||
| + | <br> | ||
| + | |||
| + | ===Characterization=== | ||
| + | <p>CAU_China 2022 used this part as their oxalate secretion circuit. We transfered this circuit into pUC18 and pBBR1MCS2 to adapt to different chassis bacteria. This phenotype, in the way we test it, is reflected in the presence of more reducing substance outside the bacteria or changes in pH.</p> | ||
| + | <br> | ||
| + | In <i>E.coli</i> DH5-alpha, we tested the gene by simple quantitive test of the generation and concentration variation of oxalic acid. This was accomplished by testing pH changes along with time in the liquid medium of the bacterial solution during 10 hours after induction. In this experiment, the three groups were separately inoculated into 10 mL liquid LB medium. The culture was induced after two hours of shaking at 37 ℃. After that, cultures were continued in the original conditions and the pH of the solution was measured every two hours. | ||
| + | |||
| + | [[File:CAU-oa-er.png|600px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b> Fig.1 Result of oxalic acid secretion in <i>E. coli</i> tested by measuring pH. | ||
| + | The origin pH is LB medium, since oxalic acid is weak acid, the efflux of oxalate will lead to the increase of pH outside bacteria. And the growth of bacteria itself will also increase the pH, so theoretically the bacteria transferred into the genetic circuit will have a higher pH increase compared with the negative control.</b></p> | ||
| + | |||
| + | In this experiment, we can see that the experimental group has a prominent ability to increase pH. This can partly be explained by the correct phenotype, as the efflux oxalate may form alkaline sodium oxalate in the LB medium environment. | ||
| + | |||
| + | In <i>P. fluorescens</i> 2P24, we used the oxalic acid content detection kit (BoxBio, Beijing), which measure oxalic acid by salicylic acid complexed iron colorimetric method, allowing us to conduct colorimetric test for oxalic acid and other reducing substances in parallel treated samples with the same OD. | ||
| + | |||
| + | We added the same amount of induced bacterial solution to each of the medium, and they were first adjusted to the same OD<sub>595</sub> with ddH<sub>2</sub>O. The control group is <i>P. fluorescens</i> 2P24 with an empty pBBR1MCS2 vector. After shaking culture at 28 ℃ for 12 hours, they were induced by IPTG. After another 8 hours, the bacterial solution was collected before we used a vacuum concentrator concentrate the products to ten times theirs original concentration. | ||
| + | |||
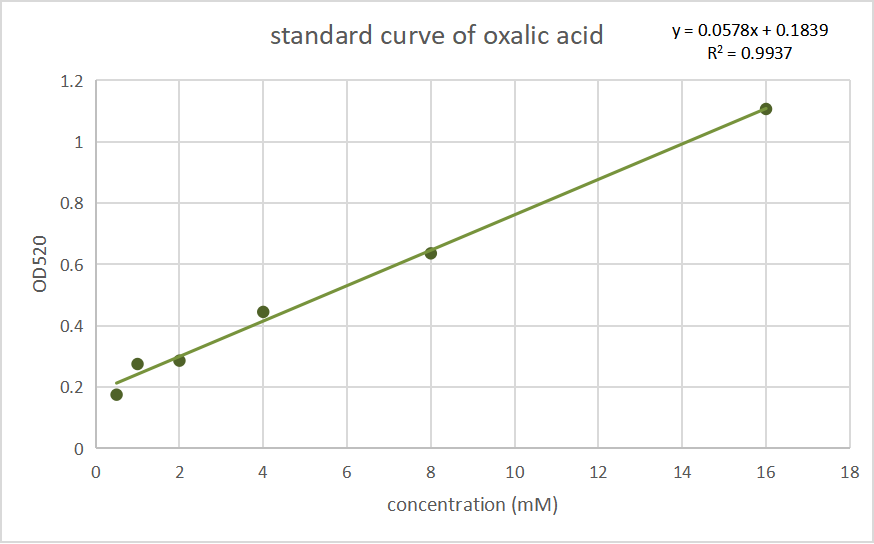
| + | [[File:Oa standard.png|600px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b>Fig.2 Standard curves of oxalate concentration and absorbance at 520 nm.</b></p> | ||
| + | |||
| + | [[File:Oakit-poaltest-2p24-pic.jpg|600px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b>Fig.3 Oxalic acid concentration test using 96-well plate. Lighter the red, higher the oxalic acid concentration in the well.</b></p> | ||
| + | |||
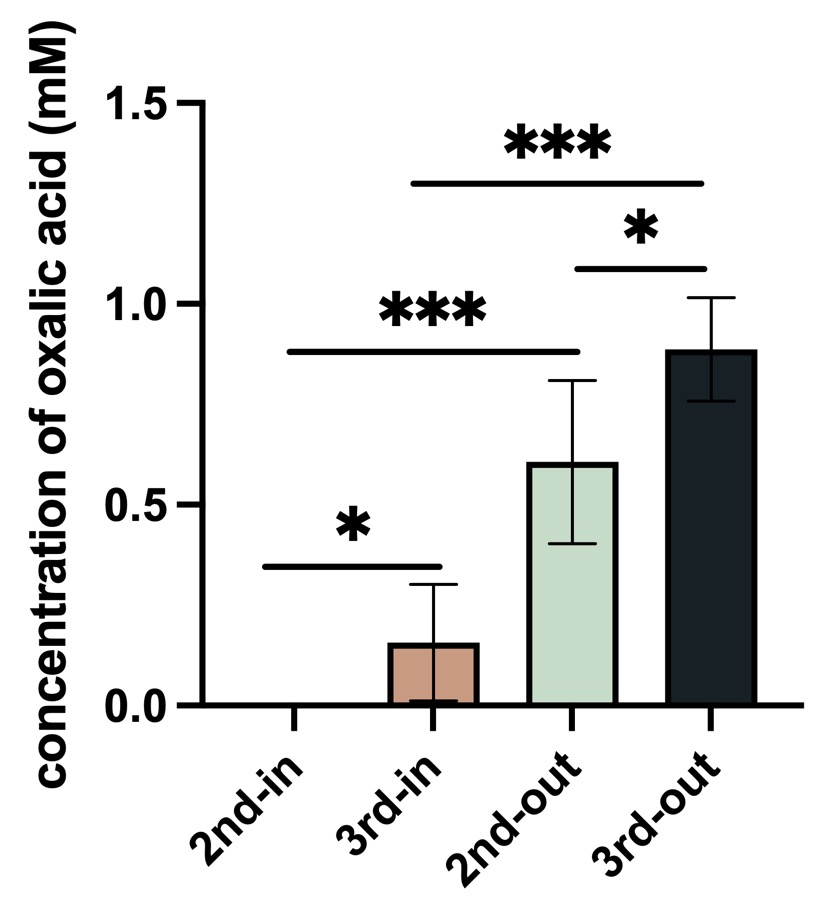
| + | [[File:CAUChinaoa2P24.jpeg|600px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b>Fig.4 Result of oxalic acid concentration in <i>P. fluorescens</i>.</b></p> | ||
| + | |||
| + | <p>Control: empty vector, 2nd: BBa_K4192120, 3rd:BBa_K4192121. In: Bacteria lysate, detected the oxalic acid concentration inside of the bacteria, out: the supernate of bacteria solution ,detected the oxalic acid concentration outside of the bacteria.</p> | ||
| + | |||
| + | <p>According to the results of ANOVA (Duncan's method), between these groups have a significant impact on the experimental results. We can come to the conclusion that the added Plac improved the efflux efficiency of oxalic acid, and this is our resulting oxalic acid producing bacteria.</p> | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | <p>This part is originated from <i>Burkholderia glumae</i> and <i>Fomitopsis palustris</i>. In our project we used this part in pBBR1MCS2 plasmid to run the tests. We tested.his part by the pH changes in the bacterial solution and by oxalic acid content detection kit (BoxBio, Beijing) to demonstrate it can produce and secrete oxalic acid in <i>E. coli</i> and <i>Pseudomonas fluorescens</i>. | ||
| + | |||
<!-- --> | <!-- --> | ||
Latest revision as of 03:12, 14 October 2022
Plac-obcA-obcB-Fpoar, Oxalate secretion gene circuit
The expression products of obcA and obcB are enzymes needed to produce oxalate from citric acid and acetyl-CoA. And FpOAR protein can efflux oxalate. The composite part of these three genes can efflux oxalic acid out of the thallus. Because it only have one promoter at the beginning of the whole genetic circuit, the efflux rate of the generated oxalic acid is relatively low, which may also lead to the death of the bacteria more easily due to excessive accumulation of oxalic acid in the body. We have already tested it in E.coli and Pseudomonas fluorescens.
Characterization
CAU_China 2022 used this part as their oxalate secretion circuit. We transfered this circuit into pUC18 and pBBR1MCS2 to adapt to different chassis bacteria. This phenotype, in the way we test it, is reflected in the presence of more reducing substance outside the bacteria or changes in pH.
In E.coli DH5-alpha, we tested the gene by simple quantitive test of the generation and concentration variation of oxalic acid. This was accomplished by testing pH changes along with time in the liquid medium of the bacterial solution during 10 hours after induction. In this experiment, the three groups were separately inoculated into 10 mL liquid LB medium. The culture was induced after two hours of shaking at 37 ℃. After that, cultures were continued in the original conditions and the pH of the solution was measured every two hours.
Fig.1 Result of oxalic acid secretion in E. coli tested by measuring pH. The origin pH is LB medium, since oxalic acid is weak acid, the efflux of oxalate will lead to the increase of pH outside bacteria. And the growth of bacteria itself will also increase the pH, so theoretically the bacteria transferred into the genetic circuit will have a higher pH increase compared with the negative control.
In this experiment, we can see that the experimental group has a prominent ability to increase pH. This can partly be explained by the correct phenotype, as the efflux oxalate may form alkaline sodium oxalate in the LB medium environment.
In P. fluorescens 2P24, we used the oxalic acid content detection kit (BoxBio, Beijing), which measure oxalic acid by salicylic acid complexed iron colorimetric method, allowing us to conduct colorimetric test for oxalic acid and other reducing substances in parallel treated samples with the same OD.
We added the same amount of induced bacterial solution to each of the medium, and they were first adjusted to the same OD595 with ddH2O. The control group is P. fluorescens 2P24 with an empty pBBR1MCS2 vector. After shaking culture at 28 ℃ for 12 hours, they were induced by IPTG. After another 8 hours, the bacterial solution was collected before we used a vacuum concentrator concentrate the products to ten times theirs original concentration.
Fig.2 Standard curves of oxalate concentration and absorbance at 520 nm.
Fig.3 Oxalic acid concentration test using 96-well plate. Lighter the red, higher the oxalic acid concentration in the well.
Fig.4 Result of oxalic acid concentration in P. fluorescens.
Control: empty vector, 2nd: BBa_K4192120, 3rd:BBa_K4192121. In: Bacteria lysate, detected the oxalic acid concentration inside of the bacteria, out: the supernate of bacteria solution ,detected the oxalic acid concentration outside of the bacteria.
According to the results of ANOVA (Duncan's method), between these groups have a significant impact on the experimental results. We can come to the conclusion that the added Plac improved the efflux efficiency of oxalic acid, and this is our resulting oxalic acid producing bacteria.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2552
Illegal PstI site found at 397
Illegal PstI site found at 2695 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2552
Illegal PstI site found at 397
Illegal PstI site found at 2695
Illegal NotI site found at 2330 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2552
Illegal BglII site found at 1541
Illegal BglII site found at 3555
Illegal XhoI site found at 304
Illegal XhoI site found at 1428
Illegal XhoI site found at 3367 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2552
Illegal PstI site found at 397
Illegal PstI site found at 2695 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2552
Illegal PstI site found at 397
Illegal PstI site found at 2695
Illegal NgoMIV site found at 389
Illegal NgoMIV site found at 720
Illegal NgoMIV site found at 748
Illegal NgoMIV site found at 784
Illegal NgoMIV site found at 1014
Illegal NgoMIV site found at 1791
Illegal NgoMIV site found at 2208
Illegal AgeI site found at 326 - 1000COMPATIBLE WITH RFC[1000]