Difference between revisions of "Part:BBa K4140006"
Ahmed Mattar (Talk | contribs) (→Usage) |
(→Usage) |
||
| (15 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
==Part Description== | ==Part Description== | ||
| − | + | ||
| + | An exclusive transporter for tyrosine is encoded by the tyrP gene in Escherichia coli. Tyrosine inhibits it, whereas phenylalanine and, to a lesser extent, tryptophan activate it. Tyrosine also inhibits its production. The TyrR protein, which has two cognate binding sites (TyrR boxes) that span nucleotides 30 to 75, mediates both of these effects when it binds to one of both of these sites. A dimer that binds to the upstream box and interacts with the subunit (CTD) of RNA polymerase causes the activation in the presence of phenylalanine or tryptophan (RNAP). A hexamer binds to both TyrR boxes during repression in the presence of tyrosine. | ||
==Usage== | ==Usage== | ||
| − | TyrP promoter | + | TyrP promoter normally regulates the expression for a part that codes for the tyrosine-specific transport system, it is induced when phenylalanine and the tyrR gene product (TyrR dimer)are together, taking advantage of that we use it to regulate the expression of PAH in our therapeutic circuit and ß-galactosidase in our diagnostic circuit as shown in figure 1. |
| + | [[File:ptyrp.png|thumb|Right|Figure 1. (shows the usage of TyrP in our circuit design) ]] | ||
| + | <br><br><br><br><br><br><br><br> | ||
==Literature Characterization== | ==Literature Characterization== | ||
| + | TyrP prompter activity is mediated by the presence of TyrR protein, In this experiment,the change of TyrP promoter activity was measured according to the nature of different DNA templates.The TyrP promoter was significantly expanded in the supercoiled form in the presence of RNAP, indicating that RNAP forms a more stable complex with the supercoiled TyrP promoter. However, only mild opening was identified at the upstream binding site's 10 areas when linear template was present. | ||
| − | [[File: | + | [[File:Tyrp-1.png|thumb|right|Figure.1 KMnO4 footprinting of linear and supercoiled templates]] |
| − | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| + | ==Characterization by mathematical modeling== | ||
| + | We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (4) and graph (1). | ||
| + | [[File:modell1.png|Right|]] | ||
| + | <br> | ||
| + | Figure (4) represents the cascade of reactions in whole cell-based biosensor model. | ||
| + | [[File:modell11.png|Right|]] | ||
| + | <br><br><br> | ||
| + | Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase. | ||
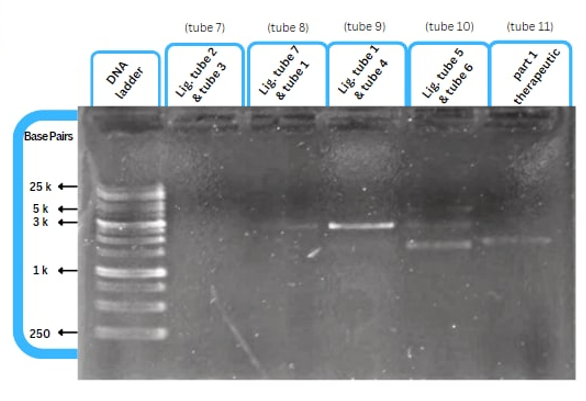
| + | ==Experimental Characterization== | ||
| + | [[File:capture7.png|right|]] | ||
| + | <br><br><br><br><br> | ||
| + | This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 6 (the last one). The running part (ordered from IDT) included T7P - TyrR RBS - TyrR - TyrPromoter. | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
==References== | ==References== | ||
| + | 1.Yang, J., Ganesan, S., Sarsero, J., and Pittard, A.J. (1993) A genetic analysis of various functions of the TyrR protein of Escherichia coli. J Bacteriol 175: 1767–1776 | ||
| + | |||
| + | 2.Yang, J., Camakaris, H., and Pittard, A.J. (1996b) In vitro transcriptional analysis of TyrR-mediated activation of the mtr and tyrP+3 promoters of Escherichia coli. J Bacteriol 178: 6389–6393 | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 18:52, 11 October 2022
TyrP promoter
Part Description
An exclusive transporter for tyrosine is encoded by the tyrP gene in Escherichia coli. Tyrosine inhibits it, whereas phenylalanine and, to a lesser extent, tryptophan activate it. Tyrosine also inhibits its production. The TyrR protein, which has two cognate binding sites (TyrR boxes) that span nucleotides 30 to 75, mediates both of these effects when it binds to one of both of these sites. A dimer that binds to the upstream box and interacts with the subunit (CTD) of RNA polymerase causes the activation in the presence of phenylalanine or tryptophan (RNAP). A hexamer binds to both TyrR boxes during repression in the presence of tyrosine.
Usage
TyrP promoter normally regulates the expression for a part that codes for the tyrosine-specific transport system, it is induced when phenylalanine and the tyrR gene product (TyrR dimer)are together, taking advantage of that we use it to regulate the expression of PAH in our therapeutic circuit and ß-galactosidase in our diagnostic circuit as shown in figure 1.
Literature Characterization
TyrP prompter activity is mediated by the presence of TyrR protein, In this experiment,the change of TyrP promoter activity was measured according to the nature of different DNA templates.The TyrP promoter was significantly expanded in the supercoiled form in the presence of RNAP, indicating that RNAP forms a more stable complex with the supercoiled TyrP promoter. However, only mild opening was identified at the upstream binding site's 10 areas when linear template was present.
Characterization by mathematical modeling
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (4) and graph (1).
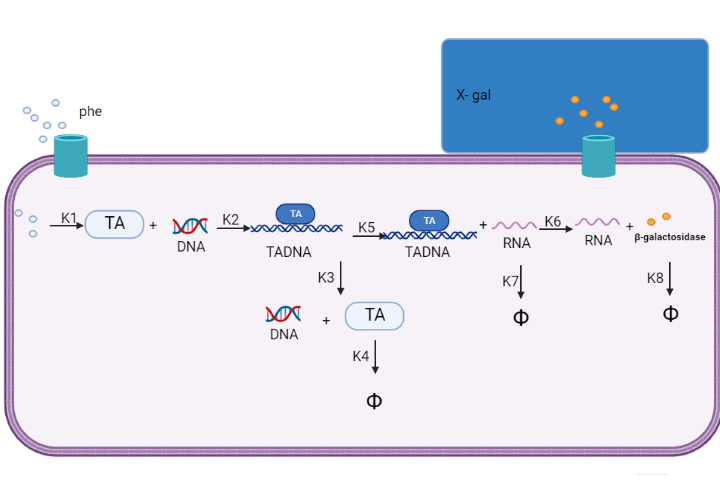
Figure (4) represents the cascade of reactions in whole cell-based biosensor model.
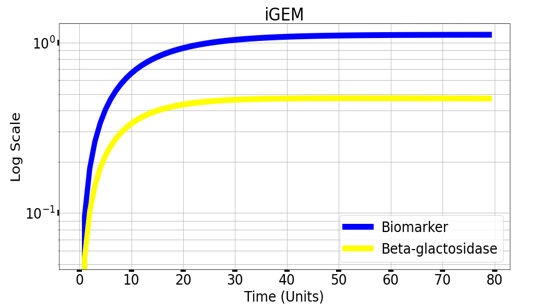
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase.
Experimental Characterization
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 6 (the last one). The running part (ordered from IDT) included T7P - TyrR RBS - TyrR - TyrPromoter.
References
1.Yang, J., Ganesan, S., Sarsero, J., and Pittard, A.J. (1993) A genetic analysis of various functions of the TyrR protein of Escherichia coli. J Bacteriol 175: 1767–1776
2.Yang, J., Camakaris, H., and Pittard, A.J. (1996b) In vitro transcriptional analysis of TyrR-mediated activation of the mtr and tyrP+3 promoters of Escherichia coli. J Bacteriol 178: 6389–6393
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]