Difference between revisions of "Part:BBa K1529150"
Alexandra L (Talk | contribs) (→Characterization) |
Alexandra L (Talk | contribs) (→Contribution From CAU_China 2022) |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 19: | Line 19: | ||
<br> | <br> | ||
===Characterization=== | ===Characterization=== | ||
| − | <p>CAU_China 2022 used the CDS sequence in their oxalate secretion | + | <p>CAU_China 2022 used the CDS sequence in their oxalate secretion circuit and the expression product of <i>obcA</i> is one of the main enzyme in this circuit. It can act together with ObcB to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA)<sup>[1]</sup>. When there is only ObcA, it has a low ability to form oxalic acid in our project.</p> |
<p>Hence, we aim to verify that the gene <i>obcA</i>, originally from <i> Burkholderia glumae</i>, can be regulated by PT7 and lac operator and properly expressed downstream to produce oxalic acid in <i>Escherichia coli</i>.</p> | <p>Hence, we aim to verify that the gene <i>obcA</i>, originally from <i> Burkholderia glumae</i>, can be regulated by PT7 and lac operator and properly expressed downstream to produce oxalic acid in <i>Escherichia coli</i>.</p> | ||
<br> | <br> | ||
[[File:PathA30.png|600px|thumb|center|]] | [[File:PathA30.png|600px|thumb|center|]] | ||
| − | <p style="text-align: center;"><b>Fig.1 Genetic circuit for | + | <p style="text-align: center;"><b>Fig.1 Genetic circuit for <i>obcA</i> verification.</b></p> |
| − | |||
<p>We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). And for the T7 promoter, T7 terminator and lac operator, we just followed the structure that came with pET30a.</p> | <p>We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). And for the T7 promoter, T7 terminator and lac operator, we just followed the structure that came with pET30a.</p> | ||
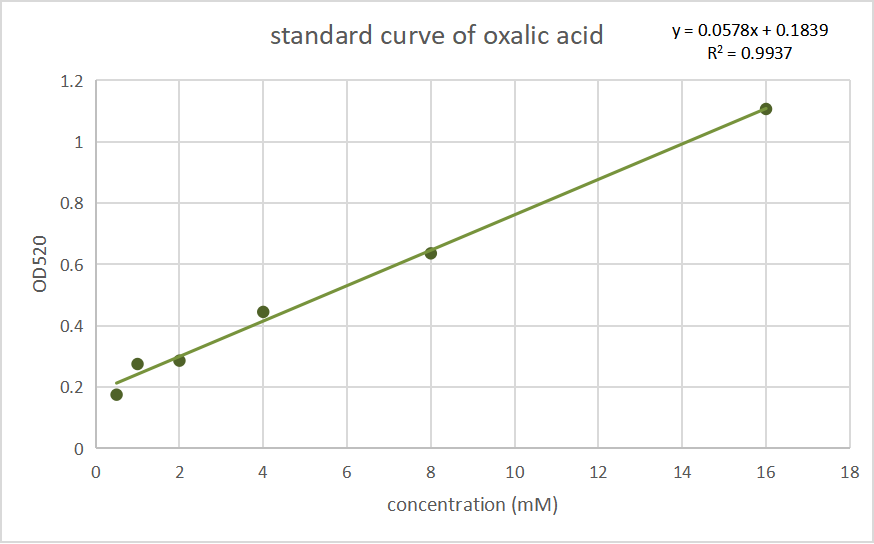
| − | <p>After sequencing and amplifying, that vector as well as unmodified pET30a were transferred into <i>E.coli</i> BL21 (DE3) separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 200 rpm, 37 ℃ for 12 h, the OD< | + | <p>After sequencing and amplifying, that vector as well as unmodified pET30a were transferred into <i>E.coli</i> BL21 (DE3) separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 200 rpm, 37 ℃ for 12 h, the OD<sub>600</sub> of the two bacteria were adjusted to nearly the same (nearly 1.3) using ddH<sub>2</sub>O, then they were induced by IPTG. Before induction, half of the two bacterial solutions were divided into the control group without induction, which were use as a blank control for colorimetric method. After a cultivation at 120 rpm, 28 ℃ for 8 h, the bacterial solution was collected and then centrifuged at 8000 xg for 1 min. The precipitate was resuspended in a ratio of 200 μl of water per 3 ml of product of the bacterial solution, and ultrasonic crushing for 10 minutes, according to the ultrasonic 3 seconds pause 3 seconds, Amp1 30% parameters. we measured its oxalic acid concentration by Oxalic Acid Content Detection Kit (BOXBIO, Beijing, China), a kit for the determination of oxalic acid concentration by salicylic acid iron colorimetric method. We tested the absorbance at OD<sub>520</sub> and convert it into the concentration of oxalic acid inside the thallus. Data are shown below:</p> |
<br> | <br> | ||
| + | [[File:Oa standard.png|300px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b>Fig.2 Standard curves of oxalate concentration and absorbance at 520 nm.</b></p> | ||
| − | </b></p> | + | [[File:CAU-A30-result.JPG|600px|thumb|center|]] |
| + | <p style="text-align: center;"><b>Fig.3 Oxalic acid concentration results in the verification of <i>obcA</i>.</b></p> | ||
| + | <p>The control is negative control, which is an empty pET30 vector. According to the result of t-test(p (0.003) < 0.05), there is significant difference between the two groups of data, indicating that <i>obcA</i> has a great influence on the the oxalic acid produced by the bacteria. </p> | ||
| + | |||
| + | <p>We finally come to the conclusion that <i>obcA</i> can respond to salinity changes in <i>E.coli</i>. Another two parallel experiments were done later, which supported our conclusion. Unfortunately, this concentration is not as high as expected, but it should act with <i>obcB</i> to produce more oxalic acid, as suggested in the literature.<sup>[2]</sup></p> | ||
| + | |||
| + | <br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1529150 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1529150 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ===References=== | ||
| + | <p>[1]Oh, Juntaek et al. “Structural basis for bacterial quorum sensing-mediated oxalogenesis.” The Journal of biological chemistry vol. 289,16 (2014): 11465-11475. doi:10.1074/jbc.M113.543462</p> | ||
| + | <p>[2]Nakata, Paul A, and Cixin He. “Oxalic acid biosynthesis is encoded by an operon in Burkholderia glumae.” FEMS microbiology letters vol. 304,2 (2010): 177-82. doi:10.1111/j.1574-6968.2010.01895.x</p> | ||
Latest revision as of 13:16, 9 October 2022
PT7_obcA
1
Contribution From CAU_China 2022
Group: CAU_China, 2022 https://2022.igem.org/Team:CAU_China
Author: Liu Muhua
Summary: Verified that this composite Part can produce oxalic acid in Escherichia coli
Characterization
CAU_China 2022 used the CDS sequence in their oxalate secretion circuit and the expression product of obcA is one of the main enzyme in this circuit. It can act together with ObcB to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA)[1]. When there is only ObcA, it has a low ability to form oxalic acid in our project.
Hence, we aim to verify that the gene obcA, originally from Burkholderia glumae, can be regulated by PT7 and lac operator and properly expressed downstream to produce oxalic acid in Escherichia coli.
Fig.1 Genetic circuit for obcA verification.
We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). And for the T7 promoter, T7 terminator and lac operator, we just followed the structure that came with pET30a.
After sequencing and amplifying, that vector as well as unmodified pET30a were transferred into E.coli BL21 (DE3) separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 200 rpm, 37 ℃ for 12 h, the OD600 of the two bacteria were adjusted to nearly the same (nearly 1.3) using ddH2O, then they were induced by IPTG. Before induction, half of the two bacterial solutions were divided into the control group without induction, which were use as a blank control for colorimetric method. After a cultivation at 120 rpm, 28 ℃ for 8 h, the bacterial solution was collected and then centrifuged at 8000 xg for 1 min. The precipitate was resuspended in a ratio of 200 μl of water per 3 ml of product of the bacterial solution, and ultrasonic crushing for 10 minutes, according to the ultrasonic 3 seconds pause 3 seconds, Amp1 30% parameters. we measured its oxalic acid concentration by Oxalic Acid Content Detection Kit (BOXBIO, Beijing, China), a kit for the determination of oxalic acid concentration by salicylic acid iron colorimetric method. We tested the absorbance at OD520 and convert it into the concentration of oxalic acid inside the thallus. Data are shown below:
Fig.2 Standard curves of oxalate concentration and absorbance at 520 nm.
Fig.3 Oxalic acid concentration results in the verification of obcA.
The control is negative control, which is an empty pET30 vector. According to the result of t-test(p (0.003) < 0.05), there is significant difference between the two groups of data, indicating that obcA has a great influence on the the oxalic acid produced by the bacteria.
We finally come to the conclusion that obcA can respond to salinity changes in E.coli. Another two parallel experiments were done later, which supported our conclusion. Unfortunately, this concentration is not as high as expected, but it should act with obcB to produce more oxalic acid, as suggested in the literature.[2]
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 276
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 276
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1420
Illegal XhoI site found at 183
Illegal XhoI site found at 1307
Illegal XhoI site found at 1751 - 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 276
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 276
Illegal NgoMIV site found at 268
Illegal NgoMIV site found at 599
Illegal NgoMIV site found at 627
Illegal NgoMIV site found at 663
Illegal NgoMIV site found at 893
Illegal NgoMIV site found at 1670
Illegal NgoMIV site found at 1721
Illegal AgeI site found at 205 - 1000COMPATIBLE WITH RFC[1000]
References
[1]Oh, Juntaek et al. “Structural basis for bacterial quorum sensing-mediated oxalogenesis.” The Journal of biological chemistry vol. 289,16 (2014): 11465-11475. doi:10.1074/jbc.M113.543462
[2]Nakata, Paul A, and Cixin He. “Oxalic acid biosynthesis is encoded by an operon in Burkholderia glumae.” FEMS microbiology letters vol. 304,2 (2010): 177-82. doi:10.1111/j.1574-6968.2010.01895.x