Difference between revisions of "Part:BBa K3861005"
Martje1998 (Talk | contribs) |
|||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
==='''Introduction'''=== | ==='''Introduction'''=== | ||
<html> | <html> | ||
| − | iGEM Humboldt_Berlin 2021 standard backbone | + | iGEM Humboldt_Berlin 2021 standard backbone derived from pSB1C3. It contains the standard high-copy-number ColE1/pMB1/pUC origin of replication as well as the standard transcriptional terminators adjacent to the BioBrick prefix and suffix. The chloramphenicol resistance cassette of pSB1C3 was replaced with the coding sequences of <I>thyA</I> (<a href="https://parts.igem.org/Part:BBa_K3861003">BBa_K3861003</a>) and <I>lldR</I> (<a href="https://parts.igem.org/Part:BBa_K3861004">BBa_K3861004</a>). The expression of <I>thyA</I> and <I>lldR</I> is driven by the synthetic constitutive promoter P_syn-thyA (<a href="https://parts.igem.org/Part:BBa_K3861002">BBa_K3861002</a>) (Stringer et al., 2013). pKF01 still contains the binding sites for the standard sequencing/amplification primers for BioBrick parts, VF2 and VF. |
| + | |||
</html> | </html> | ||
| − | ==='''Biology'''=== | + | ==='''Usage and Biology'''=== |
<html> | <html> | ||
| + | |||
| + | This backbone was used for the characterisation of the PlldR promoter (<a href="https://parts.igem.org/Part:BBa_K1847008">BBa_K1847008</a>) with the GFP reporter (<a href="https://parts.igem.org/Part:BBa_E0840">BBa_E0840</a>) in <I>Salmonella</I> Typhimurium. | ||
| + | This backbone is intended to be used in the composite part <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K3861026">BBa_K3861026</a>. | ||
| + | |||
| + | We ordered the sequence with the RCF25 BioBrick suffix and prefix sequences at the termini, double digested the gBlock with AgeI and NgoMIV. This allowed for circularization of the plasmid. | ||
| + | Subsequently, we transformed the plasmid into a <I>thyA</I> deletion <I>Salmonella</I> Typhimurium background. This mutation is required since the complementation of <I>thyA</I> acts as maintenance marker to keep the plasmid in the bacteria. After this, the growth medium of strain background no longer needed to be supplement with thymine. | ||
| + | We were also able to isolate the plasmid from this strain in order to sequence the plasmid. The sequencing showed correct results. | ||
| + | |||
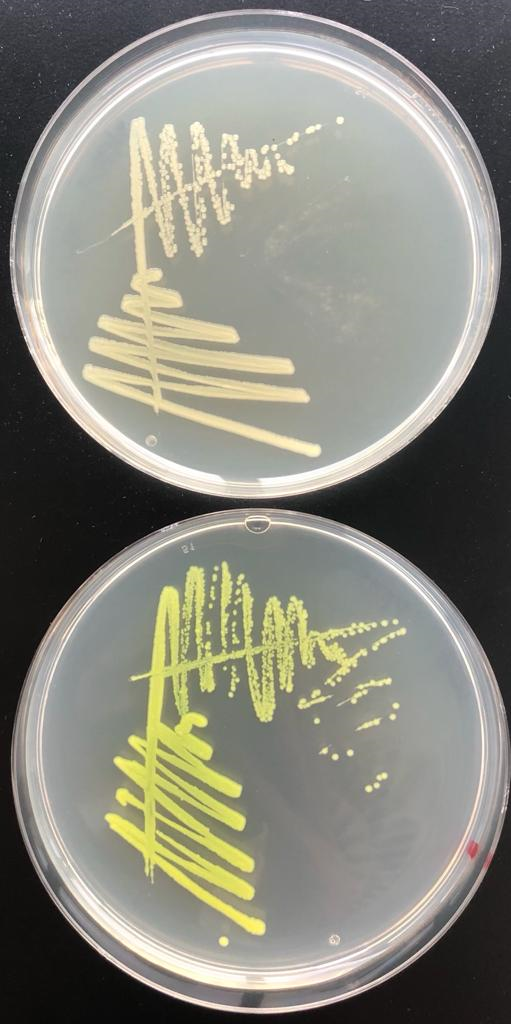
| + | Furthermore, the PlldR promoter was repressed by LldR, encoded on the plasmid. This was visible already on agar plates since the PlldR-GFP part inserted in pSB1C3 backbone led to green colored colonies, whereas this part inserted in pKF01 showed almost colorless colonies (Fig. 1). | ||
| + | |||
| + | |||
| + | |||
<figure> | <figure> | ||
| − | < | + | <img src="https://2021.igem.org/wiki/images/a/a9/T--Humboldt_Berlin--pKF01.png" alt="pKF01" width="350"> |
| − | <figcaption>GFP | + | <figcaption>Fig. 1: Coloring of <I>Salmonella</I> Typhimurium colonies harboring either PlldR-GFP inserted in pKF01 vector (plate on top) or in pSB1C3 vector (plate below).</figcaption> |
</figure> | </figure> | ||
</html> | </html> | ||
| Line 19: | Line 32: | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<!-- --> | <!-- --> | ||
| − | + | ===Sequence and Features=== | |
<partinfo>BBa_K3861005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3861005 SequenceAndFeatures</partinfo> | ||
Latest revision as of 22:22, 21 October 2021
pKF01
Introduction
iGEM Humboldt_Berlin 2021 standard backbone derived from pSB1C3. It contains the standard high-copy-number ColE1/pMB1/pUC origin of replication as well as the standard transcriptional terminators adjacent to the BioBrick prefix and suffix. The chloramphenicol resistance cassette of pSB1C3 was replaced with the coding sequences of thyA (BBa_K3861003) and lldR (BBa_K3861004). The expression of thyA and lldR is driven by the synthetic constitutive promoter P_syn-thyA (BBa_K3861002) (Stringer et al., 2013). pKF01 still contains the binding sites for the standard sequencing/amplification primers for BioBrick parts, VF2 and VF.
Usage and Biology
This backbone was used for the characterisation of the PlldR promoter (BBa_K1847008) with the GFP reporter (BBa_E0840) in Salmonella Typhimurium.
This backbone is intended to be used in the composite part BBa_K3861026.
We ordered the sequence with the RCF25 BioBrick suffix and prefix sequences at the termini, double digested the gBlock with AgeI and NgoMIV. This allowed for circularization of the plasmid.
Subsequently, we transformed the plasmid into a thyA deletion Salmonella Typhimurium background. This mutation is required since the complementation of thyA acts as maintenance marker to keep the plasmid in the bacteria. After this, the growth medium of strain background no longer needed to be supplement with thymine.
We were also able to isolate the plasmid from this strain in order to sequence the plasmid. The sequencing showed correct results.
Furthermore, the PlldR promoter was repressed by LldR, encoded on the plasmid. This was visible already on agar plates since the PlldR-GFP part inserted in pSB1C3 backbone led to green colored colonies, whereas this part inserted in pKF01 showed almost colorless colonies (Fig. 1).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 997
Illegal XhoI site found at 2804 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
