Difference between revisions of "Part:BBa K3809010"
| (2 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
To verify that ESR1 would fold correctly and there would be no steric obstacle, we simulated the protein with its corresponding additions using the tool Robetta (https://robetta.bakerlab.org/). When we recieved the model, we made an structural alignment using the natural protein as reference with a PDB code 2IOG (Protein Data Bank, rcsb.org). Therefore, we concluded that the addition of the Linker, His Tag and Signal Peptide were not interfering with the correct folding of ESR1. Furthermore, we proved that the His Tag and the linker were exposed, so we could use them for the purposed we established previously. | To verify that ESR1 would fold correctly and there would be no steric obstacle, we simulated the protein with its corresponding additions using the tool Robetta (https://robetta.bakerlab.org/). When we recieved the model, we made an structural alignment using the natural protein as reference with a PDB code 2IOG (Protein Data Bank, rcsb.org). Therefore, we concluded that the addition of the Linker, His Tag and Signal Peptide were not interfering with the correct folding of ESR1. Furthermore, we proved that the His Tag and the linker were exposed, so we could use them for the purposed we established previously. | ||
| + | <html> | ||
<div class=WordSection1> | <div class=WordSection1> | ||
<p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | <p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | ||
107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | 107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| − | color:black;mso-ansi-language:EN-US'></span></b></p> | + | color:black;mso-ansi-language:EN-US'><o:p> </o:p></span></b></p> |
<p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | <p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | ||
107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | 107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| − | color:black;mso-ansi-language:EN-US'> | + | color:black;mso-ansi-language:EN-US'>Biology and characterization of |
| + | BBa_K3809010<o:p></o:p></span></b></p> | ||
<p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| − | color:black;mso-ansi-language:EN-US'> | + | color:black;mso-ansi-language:EN-US'>We wanted to test if our protein receptor |
| − | + | would fold adequately. However, before we could enter the lab, we could not do | |
| − | the | + | it experimentally, so we recurred to an <span class=GramE><i>in silico</i></span> |
| − | + | analysis. We wanted to verify that all the modifications and features we added wouldn’t | |
| − | class=SpellE> | + | interfere with the correct folding of BBa_K3809010; furthermore, we wanted to |
| − | the | + | verify that the His Tag we <span class=GramE>added</span> and the Linker |
| + | sequence would be exposed enough for us to perform an efficient purification | ||
| + | and an adequate immobilization. We tested this through the prediction of a | ||
| + | protein model made using the Software <span class=SpellE>Robetta</span> [1]. | ||
| + | After we took the best model <span class=SpellE>Robetta</span> could offer, we | ||
| + | performed a Structural Alignment using the Human Estrogen Receptor Alpha 2IOG from | ||
| + | PDB [2] as reference. Our results confirmed that the folding of the protein wasn’t | ||
| + | being affected at all. <o:p></o:p></span></p> | ||
| − | <p class=MsoNormal align=center style='text-align:center'>< | + | <p class=MsoNormal align=center style='text-align:center'><span |
| − | + | style='font-family:"Arial",sans-serif;color:black;mso-no-proof:yes'> | |
| − | color: | + | <img width=344 height=334 |
| − | src="https://2021.igem.org/wiki/images/ | + | src="https://2021.igem.org/wiki/images/1/18/T--TecCEM--PartsRegister3image002.jpg" |
| − | alt="Imagen que contiene | + | alt="Imagen que contiene luz Descripción generada automáticamente" |
| − | v:shapes=" | + | v:shapes="Imagen_x0020_1"><![endif]></span><span lang=EN-US style='font-size: |
| − | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | + | 10.0pt;line-height:107%;font-family:"Arial",sans-serif;mso-fareast-font-family: |
| − | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'></span></p> | + | "Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> |
<p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
| Line 42: | Line 51: | ||
1.</span></b><span lang=EN-US style='font-size:10.0pt;line-height:107%; | 1.</span></b><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| − | color:black;mso-ansi-language:EN-US'> | + | color:black;mso-ansi-language:EN-US'> Protein designed by <span class=SpellE>iGEM</span> |
| − | + | <span class=SpellE>TecCEM</span>. It includes the whole receptor <span | |
| − | + | class=SpellE>hER</span></span><span lang=EN-US style='font-size:10.0pt; | |
| − | + | line-height:107%;font-family:"Cambria Math",serif;mso-fareast-font-family:"Times New Roman"; | |
| + | mso-bidi-font-family:"Cambria Math";color:black;mso-ansi-language:EN-US'>⍺</span><span | ||
| + | lang=EN-US style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'> | ||
| + | (red), a linker sequence (green), a His Tag (magenta) and a Signal Peptide | ||
| + | Sequence (blue). <span class=GramE>It can be seen that both</span> the linker | ||
| + | sequence and the His Tag are exposed. Protein visualized with Chimera [3].<o:p></o:p></span></p> | ||
| − | <p class=MsoNormal align=center style='text-align:center'>< | + | <p class=MsoNormal align=center style='text-align:center'><span |
| − | + | style='font-family:"Arial",sans-serif;color:black;mso-no-proof:yes'><img width=396 height=348 | |
| − | color: | + | src="https://2021.igem.org/wiki/images/0/0c/T--TecCEM--PartsRegister3image004.jpg" |
| − | src="https://2021.igem.org/wiki/images/ | + | alt="Imagen que contiene collar, luz Descripción generada automáticamente" |
| − | alt="Imagen que contiene | + | v:shapes="Imagen_x0020_2"><![endif]></span><span lang=EN-US style='font-size: |
| − | v:shapes=" | + | 10.0pt;line-height:107%;font-family:"Arial",sans-serif;mso-fareast-font-family: |
| − | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | + | "Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> |
| − | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'></span></p> | + | |
<p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| − | + | color:black;mso-ansi-language:EN-US'>Figure 2</span></span><span | |
| − | 2 | + | style='white-space:pre-wrap'></b><span lang=EN-US style='font-size:10.0pt; |
| − | font-family:"Arial",sans-serif; | + | line-height:107%;font-family:"Arial",sans-serif;color:black;mso-ansi-language: |
| − | color:black;mso-ansi-language:EN-US'> | + | EN-US'>. Structural alignment between the protein designed by <span |
| − | + | class=SpellE>iGEM</span> <span class=SpellE>TecCEM</span> (red, magenta, blue | |
| − | + | and green) and the crystallized structure of the Human Estrogen Receptor Alpha | |
| − | + | 2IOG (white). The alignment shows a preserved structure in the Ligand Binding | |
| + | Domain of the modified ESR1. </span><span class=SpellE><span style='font-size: | ||
| + | 10.0pt;line-height:107%;font-family:"Arial",sans-serif;color:black'>Protein</span></span><span | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | color:black'> <span class=SpellE>visualized</span> <span class=SpellE>with</span> | ||
| + | <span class=SpellE>Chimera</span> [3].</span><o:p></o:p></span></p> | ||
<p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| − | font-family:"Arial",sans-serif; | + | font-family:"Arial",sans-serif;color:black;mso-ansi-language:EN-US'>We also |
| − | color:black;mso-ansi-language:EN-US'></span></p> | + | performed a Molecular Docking to verify its affinity towards a group of common |
| + | EDCs: Estradiol (the natural ligand), Benzene, Bisphenol A, Dimethyl Phthalate, | ||
| + | Ethylene Glycol, Phenol, Chloroprene and Perfluorooctanoic Acid. To prepare the | ||
| + | model for the analysis, we added hydrogens to it, improving the quality of the | ||
| + | assay. To this end, we used the tool <span class=SpellE>MolProbity</span> [4]. | ||
| + | The ligands were obtained from PubChem [5] and the Docking was conducted using | ||
| + | the Software tool <span class=SpellE>SwissDock</span> [6]. From the best result | ||
| + | of the docking, we obtained its Free Gibbs Energy and used it to calculate the | ||
| + | Association Constant. We also analyzed the type of interaction. Furthermore, we | ||
| + | evaluated the formation of hydrogen bonds between the receptor and the small | ||
| + | molecule. The results are seen below.<o:p></o:p></span></p> | ||
| − | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | + | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US |
| − | font-family:"Arial",sans-serif;mso- | + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; |
| − | color:black;mso-ansi-language:EN-US'> | + | color:black;mso-ansi-language:EN-US'>Table I.</span></b><span lang=EN-US |
| − | + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | |
| − | + | color:black;mso-ansi-language:EN-US'> Results obtained from the Docking | |
| − | + | Analysis. Estradiol (highlighted) is the natural ligand of the receptor.<o:p></o:p></span></p> | |
| − | + | ||
| − | + | ||
| − | + | ||
<p class=MsoNormal align=center style='text-align:center'><span | <p class=MsoNormal align=center style='text-align:center'><span | ||
| − | style=' | + | style='mso-no-proof:yes'><img width=576 height=448 |
| − | src="https://2021.igem.org/wiki/images/ | + | src="https://2021.igem.org/wiki/images/9/91/T--TecCEM--PartsRegister3image006.jpg" |
| + | alt="Tabla Descripción generada automáticamente" v:shapes="Imagen_x0020_6"><![endif]></span><span | ||
lang=EN-US style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | lang=EN-US style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| − | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'></span></p> | + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> |
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'>To characterize the DNA fragment of ESR1 | ||
| + | we inserted our gene in pSB1C3 and transformed it in <i>E. coli</i> DH5α. | ||
| + | To verify that the insert was cloned correctly, we extracted the plasmid from a | ||
| + | DH5α culture and performed a PCR using M13 primers. We got positive results | ||
| + | shown below. <o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b style='mso-bidi-font-weight: | ||
| + | normal'><span style='font-family:"Arial",sans-serif;color:black;mso-no-proof: | ||
| + | yes'><img width=624 height=351 | ||
| + | src="https://2021.igem.org/wiki/images/f/f8/T--TecCEM--PartsRegister3image008.jpg" | ||
| + | alt="Imagen que contiene Diagrama Descripción generada automáticamente" | ||
| + | v:shapes="Imagen_x0020_7"><![endif]></span></b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> | ||
<p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| − | + | color:black;mso-ansi-language:EN-US'>Figure 3</span></b><span lang=EN-US | |
| − | 3 | + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; |
| − | font-family:"Arial",sans-serif | + | color:black;mso-ansi-language:EN-US'>. Amplification of the ESR1 fragment to |
| − | color:black;mso-ansi-language:EN-US'> | + | verify a correct cloning in a pSB1C3 vector. <o:p></o:p></span></p> |
| − | + | ||
| − | + | ||
| − | <p class=MsoNormal><span lang=EN-US style='mso-ansi-language:EN-US; | + | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US |
| − | + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | |
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p> </o:p></span></p> | ||
| − | <p class=MsoNormal><span lang=EN-US style='mso-ansi-language:EN-US'></span></p> | + | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; |
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'><o:p> </o:p></span></p> | ||
</div> | </div> | ||
| − | + | </html> | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 16:27, 21 October 2021
ESR1 with Linker, His Tag and Signal Peptide
This sequence codes for the Human Estrogen Receptor Alpha. It was modified in order to express the ligand binding domain and the DNA binding domain. The sequence also includes a 6X Histidine Tag (for its purification), a linker sequence of 4 glycines, 1 serine and 1 cistein (for its immobilization) and a signal peptide sequence (NPS4). This protein is a natural ocurring receptor found in human cells that acts as an enhacer protein which activates after its binding with its ligand. ESR1's natural ligand is Estradiol and after the molecule binds in its domain, it will undergo structural changes that activate the receptor by the formation of a dimer. It has been established that the most important aminoacids for the correct binding of estradiol and ESR1 are Gly521, His524, Leu525 and Met528.
For the purification of ESR1, we added a 6X His Tag and for the immobilization of the protein in a piezoelectric sensor, we added a linker which includes a sequence of 4 glycines, 1 serine and 1 cistein. The addition of the cistein is very important since the immobilization method we chose is based on the formation of a disulfide bond. Furthermore, we chose to mantain the DNA binding domain because it has a region rich in cisteins. Finally, we added a signal peptide sequence NPS4 (which comes from BBa_K3606042), in order to guarantee the secretion of our protein and make it easier for us to purify.
To verify that ESR1 would fold correctly and there would be no steric obstacle, we simulated the protein with its corresponding additions using the tool Robetta (https://robetta.bakerlab.org/). When we recieved the model, we made an structural alignment using the natural protein as reference with a PDB code 2IOG (Protein Data Bank, rcsb.org). Therefore, we concluded that the addition of the Linker, His Tag and Signal Peptide were not interfering with the correct folding of ESR1. Furthermore, we proved that the His Tag and the linker were exposed, so we could use them for the purposed we established previously.
Biology and characterization of
BBa_K3809010
We wanted to test if our protein receptor
would fold adequately. However, before we could enter the lab, we could not do
it experimentally, so we recurred to an in silico
analysis. We wanted to verify that all the modifications and features we added wouldn’t
interfere with the correct folding of BBa_K3809010; furthermore, we wanted to
verify that the His Tag we added and the Linker
sequence would be exposed enough for us to perform an efficient purification
and an adequate immobilization. We tested this through the prediction of a
protein model made using the Software Robetta [1].
After we took the best model Robetta could offer, we
performed a Structural Alignment using the Human Estrogen Receptor Alpha 2IOG from
PDB [2] as reference. Our results confirmed that the folding of the protein wasn’t
being affected at all.
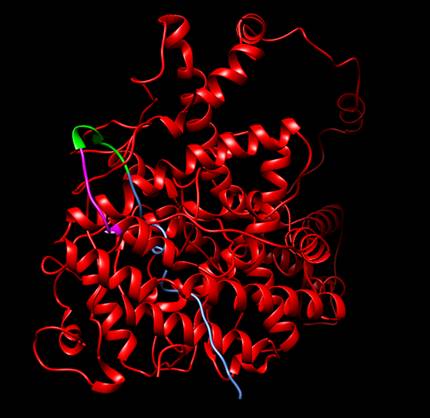
Figure
1. Protein designed by iGEM
TecCEM. It includes the whole receptor hER⍺
(red), a linker sequence (green), a His Tag (magenta) and a Signal Peptide
Sequence (blue). It can be seen that both the linker
sequence and the His Tag are exposed. Protein visualized with Chimera [3].
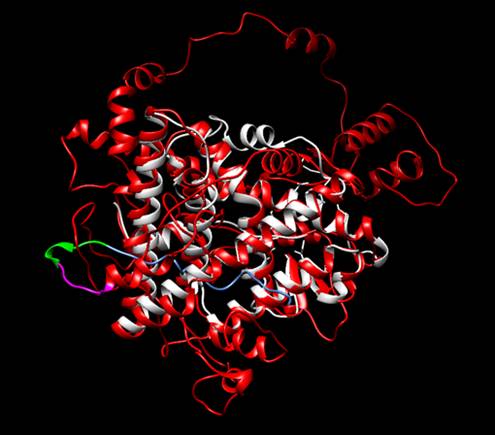
Figure 2. Structural alignment between the protein designed by iGEM TecCEM (red, magenta, blue
and green) and the crystallized structure of the Human Estrogen Receptor Alpha
2IOG (white). The alignment shows a preserved structure in the Ligand Binding
Domain of the modified ESR1. Protein visualized with
Chimera [3].
We also
performed a Molecular Docking to verify its affinity towards a group of common
EDCs: Estradiol (the natural ligand), Benzene, Bisphenol A, Dimethyl Phthalate,
Ethylene Glycol, Phenol, Chloroprene and Perfluorooctanoic Acid. To prepare the
model for the analysis, we added hydrogens to it, improving the quality of the
assay. To this end, we used the tool MolProbity [4].
The ligands were obtained from PubChem [5] and the Docking was conducted using
the Software tool SwissDock [6]. From the best result
of the docking, we obtained its Free Gibbs Energy and used it to calculate the
Association Constant. We also analyzed the type of interaction. Furthermore, we
evaluated the formation of hydrogen bonds between the receptor and the small
molecule. The results are seen below.
Table I. Results obtained from the Docking
Analysis. Estradiol (highlighted) is the natural ligand of the receptor.

To characterize the DNA fragment of ESR1
we inserted our gene in pSB1C3 and transformed it in E. coli DH5α.
To verify that the insert was cloned correctly, we extracted the plasmid from a
DH5α culture and performed a PCR using M13 primers. We got positive results
shown below.
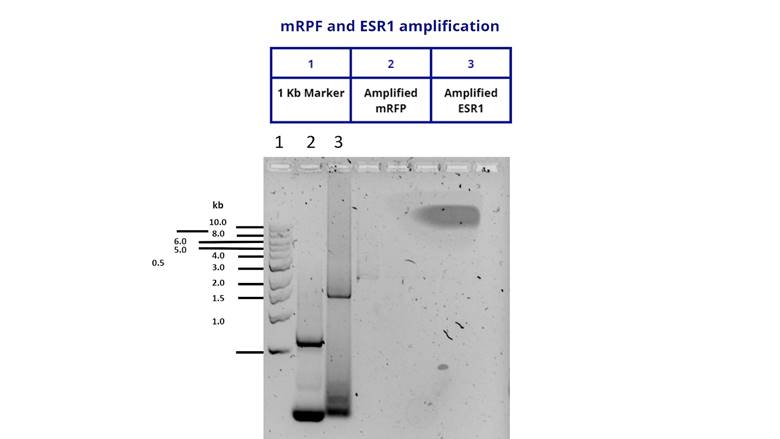
Figure 3. Amplification of the ESR1 fragment to
verify a correct cloning in a pSB1C3 vector.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 48
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1264
