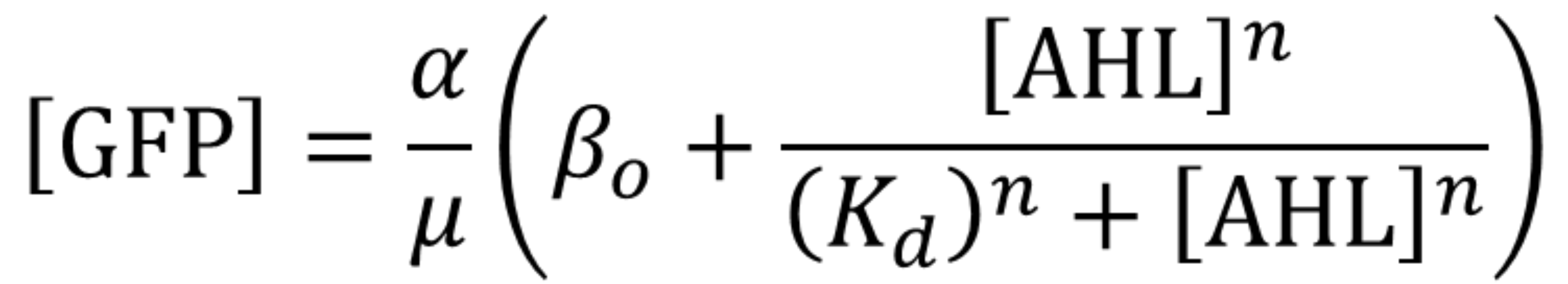Difference between revisions of "Part:BBa K2656003"
JeanHerdoiza (Talk | contribs) |
JeanHerdoiza (Talk | contribs) |
||
| Line 28: | Line 28: | ||
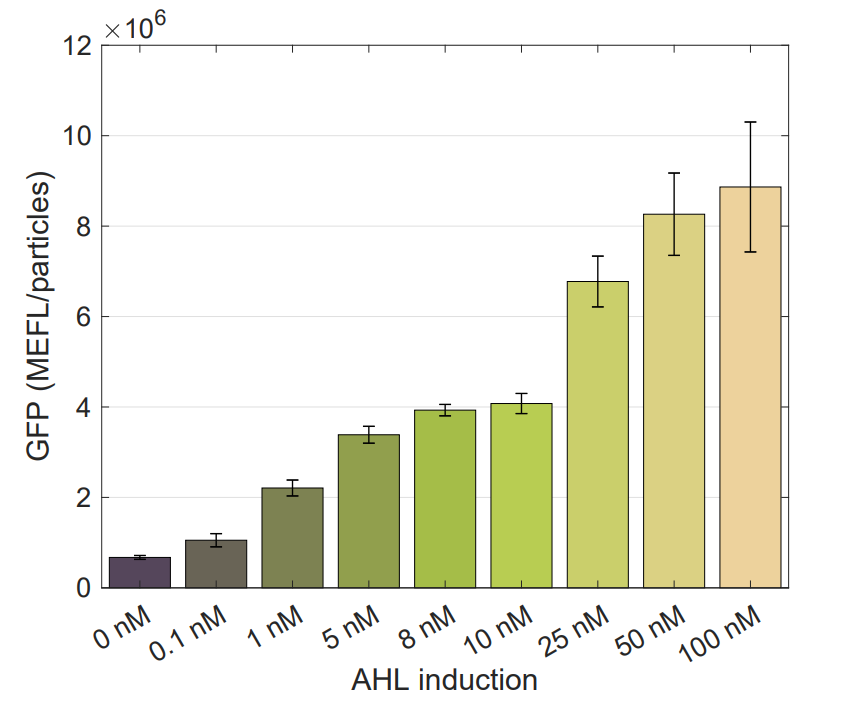
[[File:T--Ecuador--K3893028_static_response.png|600px|thumb|center|alt=domestication.|Final GFP expression levels for different AHL inducer concentrations.]] | [[File:T--Ecuador--K3893028_static_response.png|600px|thumb|center|alt=domestication.|Final GFP expression levels for different AHL inducer concentrations.]] | ||
| − | We used this part to characterize the Golden Braid version ([https://parts.igem.org/Part:BBa_K2656003 K2656003]) of pLux promoter ([https://parts.igem.org/Part:BBa_R0062 R0062]) made by [http://2018.igem.org/Team:Valencia_UPV Valencia_UPV 2018 Team]. Check our experimental page for more details. | + | |
| + | We used this part to characterize the Golden Braid version ([https://parts.igem.org/Part:BBa_K2656003 K2656003]) of pLux promoter ([https://parts.igem.org/Part:BBa_R0062 R0062]) made by [http://2018.igem.org/Team:Valencia_UPV Valencia_UPV 2018 Team]. Check our experimental page for more details, [https://2021.igem.org/File:T--Ecuador--Exp_K3893028.xlsx here you can get the raw data.] | ||
Using the same [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol as Valencia_UPV team ], we have obtained the parameters for the following model: | Using the same [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol as Valencia_UPV team ], we have obtained the parameters for the following model: | ||
| Line 60: | Line 61: | ||
[[File:T--Ecuador_UPV--Model_pLux_Charact.png|600px|thumb|center|alt=Data.|Experimental data and simulation to characterize the pLux promoter]] | [[File:T--Ecuador_UPV--Model_pLux_Charact.png|600px|thumb|center|alt=Data.|Experimental data and simulation to characterize the pLux promoter]] | ||
| − | Note: This characterization data for part R0062 (K2656003) | + | Note: This characterization data for part R0062 ([https://parts.igem.org/Part:BBa_K2656003 K2656003]) needs to be interpreted in the context of the transcriptional units from where the experimental data was taken ([https://parts.igem.org/Part:BBa_K3893028 K3893028], [https://parts.igem.org/Part:BBa_K2656122 K2656122], [https://parts.igem.org/Part:BBa_K2656114 K2656114).] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 18:44, 19 October 2021
Promoter HSL-mediated luxR
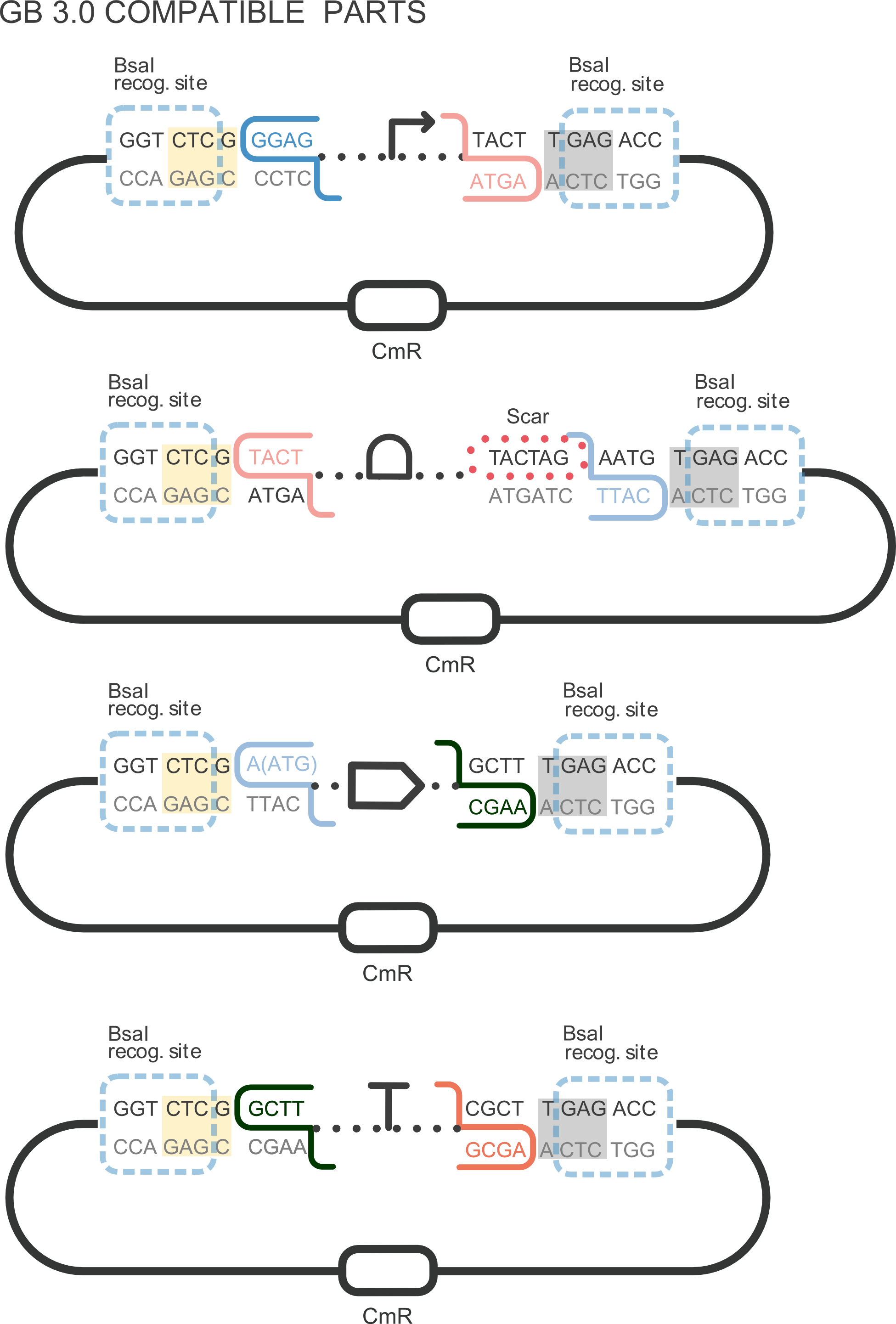
Part BBa_K2656003 is the promoter BBa_R0062 compatible with both Biobrick and [http://2018.igem.org/Team:Valencia_UPV/Design GoldenBraid 3.0] assembly methods. This promoter is positively regulated by LuxR together with Acyl-homoserine-lactone. It can be combined with other compatible parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Valencia UPV IGEM 2018 Printeria Collection] to assemble transcriptional units with the Golden Gate assembly protocol .
Ecuador 2021's characterization
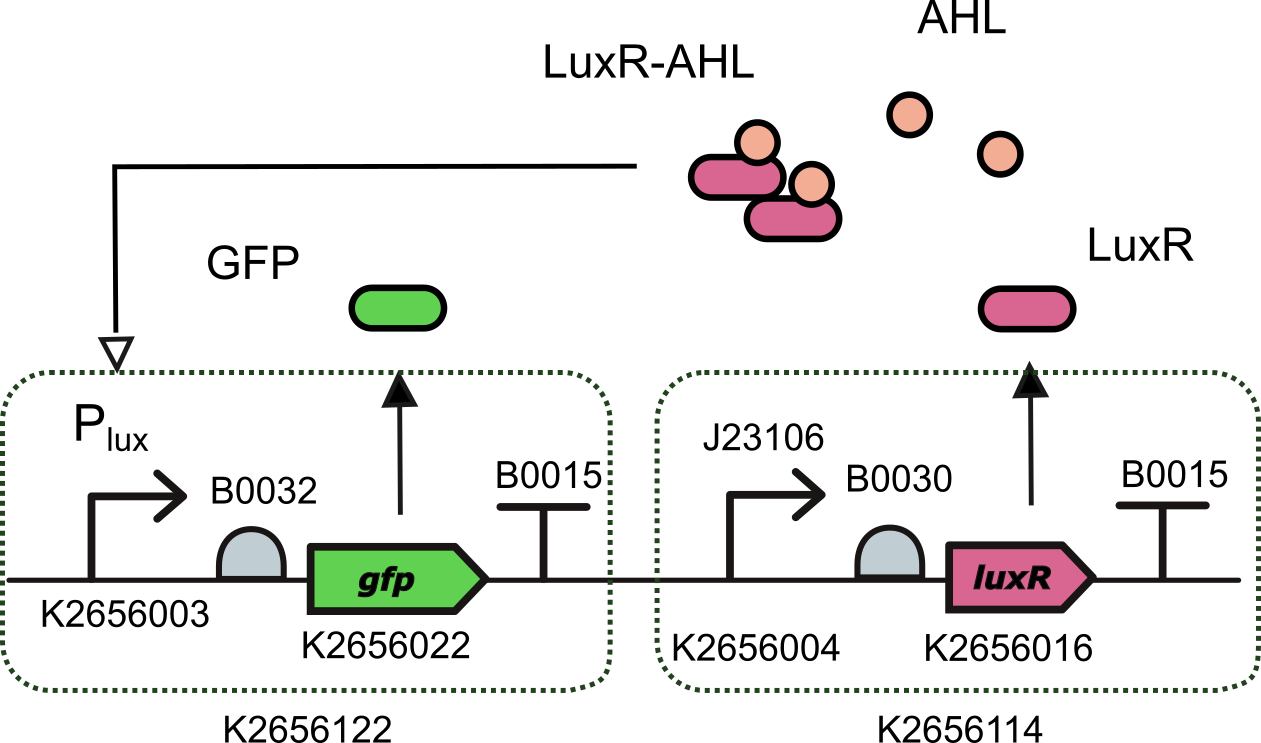
We characterized this promoter using the composite part K3893028 that we built by assembling together
- BBa_K2656122: a Level 1 transcriptional unit expresing GFP under the control of the pux promoter (Golden Braid compatible)
- BBa_K2656114: a Level 1 transcriptional unit constitutively expressing luxR gene (Golden Braid compatible)
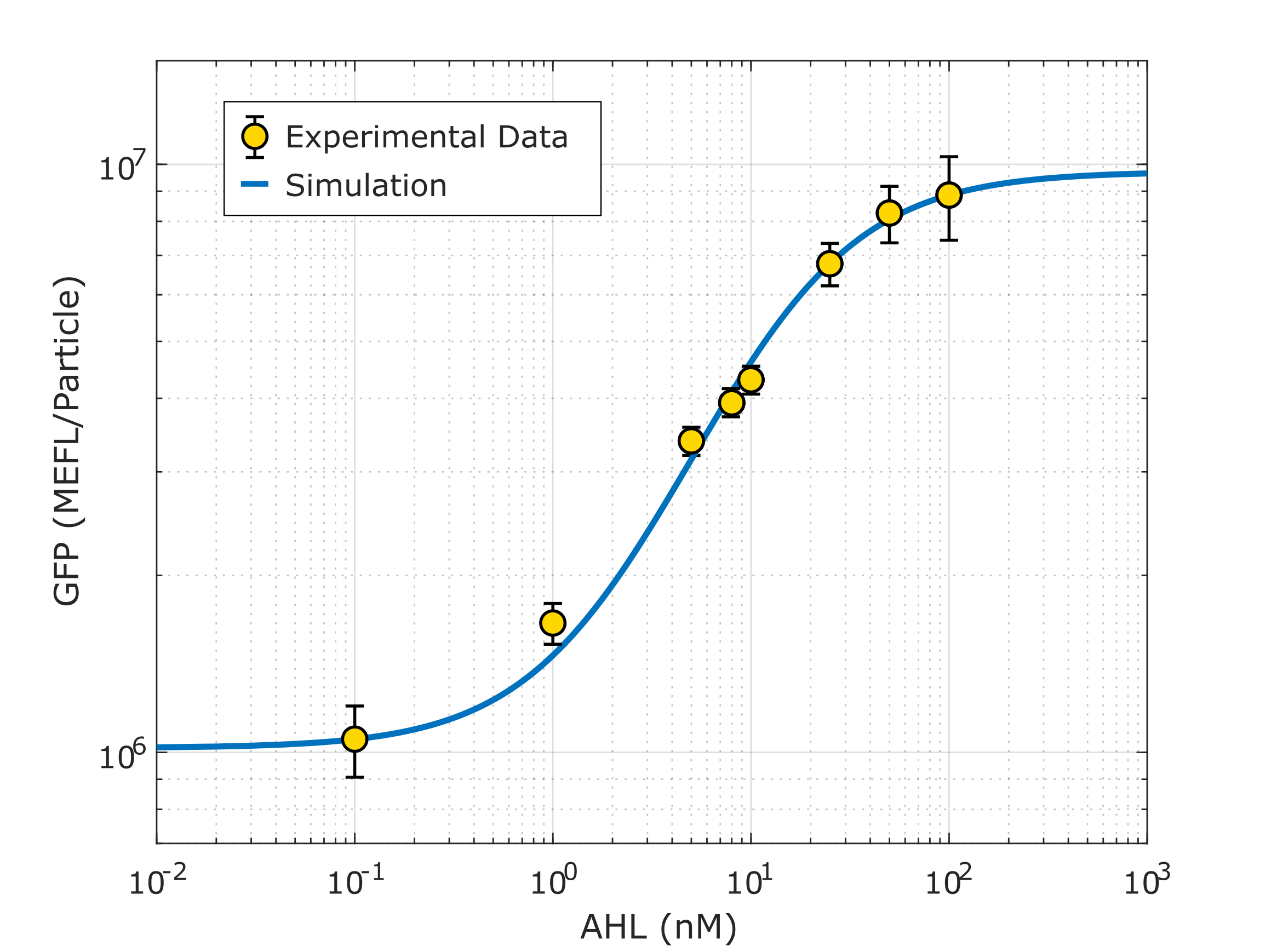
Device K3893028 allows us to measure GFP expression levels for different AHL induction concentrations.
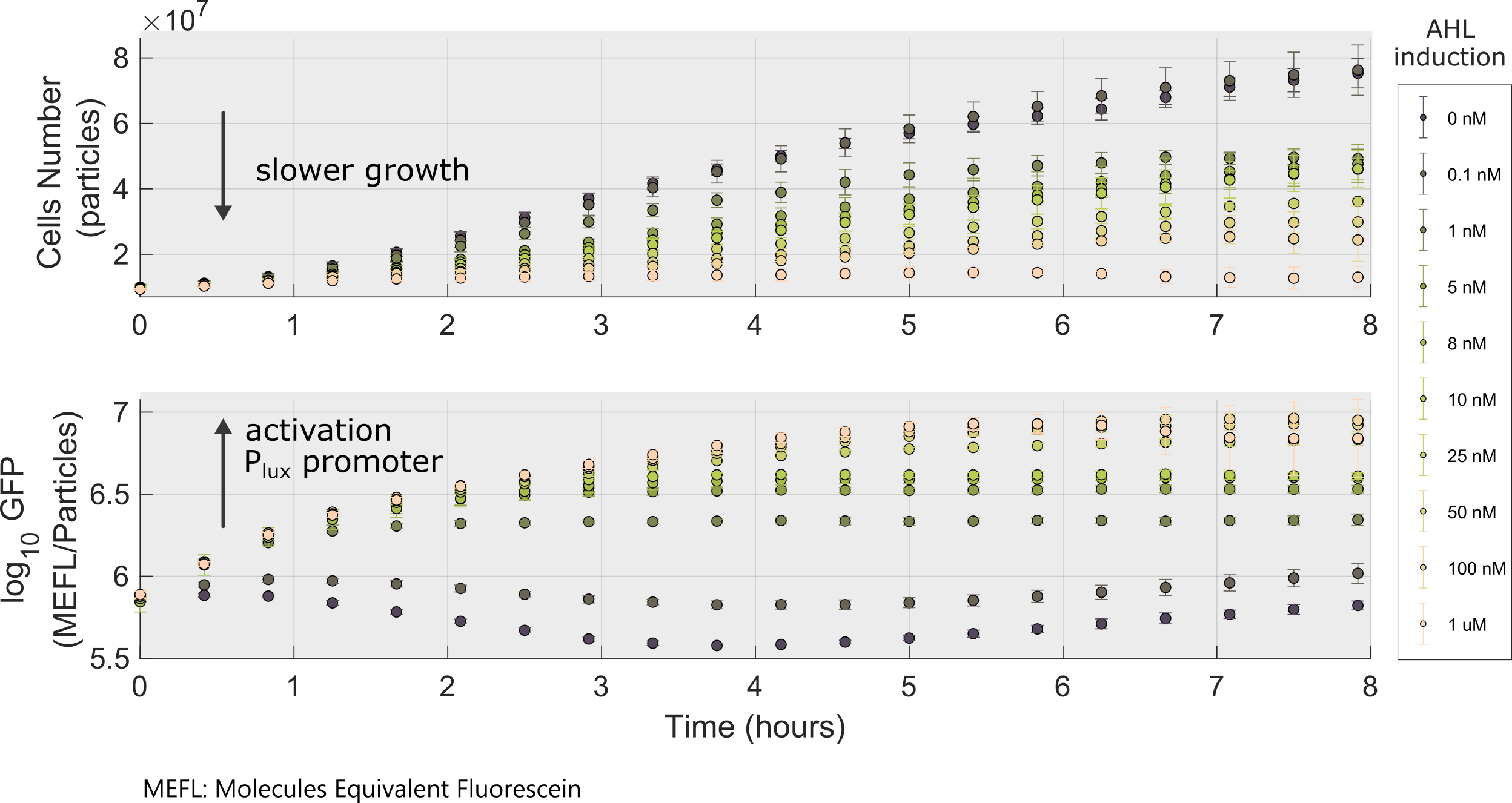
Following a standard experimental procedure and the IGEM 2018 Interlab calibration for Particles and MEFL we obtained the temporal behavior of the device for different inducer concentrations.
We used this part to characterize the Golden Braid version (K2656003) of pLux promoter (R0062) made by [http://2018.igem.org/Team:Valencia_UPV Valencia_UPV 2018 Team]. Check our experimental page for more details, here you can get the raw data.
Using the same [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol as Valencia_UPV team ], we have obtained the parameters for the following model:
| Table 1. Parameters obtained from part K3893028 corresponding to K2656003. | |||
| Parameter | Value | ||
| Effective translation rate 𝛼 | 𝛼 = 8.11e8 molec min-1 | ||
| Effective dissociation constant Kd | Kd = 15.34 nM | ||
| Hill coefficient | n = 0.958 | ||
| Basal expression | 𝛽 = 0.1049 | ||
| Dilution rate μ | μ = 0.0115 min-1 | ||
Using this optimized parameter we obtain the simulated hill function corresponding to the previous equation and compared it with the experimental data obtained (for steady-state, this is after reaching equilibrium).
Note: This characterization data for part R0062 (K2656003) needs to be interpreted in the context of the transcriptional units from where the experimental data was taken (K3893028, K2656122, K2656114).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]