Difference between revisions of "Part:BBa K3758301"
MichaelCWB (Talk | contribs) |
MichaelCWB (Talk | contribs) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 47: | Line 47: | ||
<h3>Choosing the Reporter System</h3> | <h3>Choosing the Reporter System</h3> | ||
| + | <h4>Fluorescence</h4> | ||
<p> | <p> | ||
The next important design decision was the selection of the reporter system we would use. Therefore, we looked into different fluorescent proteins, such as mScarlet and sfGFP, but also into luciferase-based reporters, like the Firefly and Nanoluc luciferase. After our first measurements, it was clear that fluorescent reporters are not suitable for our cell-free measurements in our chloroplast systems, as the signal can not be distinguished from the noise (Figure 1). On the one hand, this can be explained due to the autofluorescence of the remaining chlorophyll molecules, which increase the fluorescence background levels of the extract control. And on the other hand the expression level of our cell-free reaction seems to be too low at least for the beginning, when we were still at the troubleshooting phase of our extract preparation. | The next important design decision was the selection of the reporter system we would use. Therefore, we looked into different fluorescent proteins, such as mScarlet and sfGFP, but also into luciferase-based reporters, like the Firefly and Nanoluc luciferase. After our first measurements, it was clear that fluorescent reporters are not suitable for our cell-free measurements in our chloroplast systems, as the signal can not be distinguished from the noise (Figure 1). On the one hand, this can be explained due to the autofluorescence of the remaining chlorophyll molecules, which increase the fluorescence background levels of the extract control. And on the other hand the expression level of our cell-free reaction seems to be too low at least for the beginning, when we were still at the troubleshooting phase of our extract preparation. | ||
| Line 57: | Line 58: | ||
</figure></center></html> | </figure></center></html> | ||
| − | + | <h4>Luminescence</h4> | |
<p> | <p> | ||
Therefore, we went back to the design phase and decided to further look into luminescence-based reporters. The next experiment we performed was the comparison of the Firefly with the Nanoluc luciferase. Generally, it can be observed that the background of luminescence is very low in the control, compared to fluorescence measurements (Figure 2). Additionally, high luminescence levels can be observed, which allows the detection of successful expression from our chloroplast cell-free extracts. When comparing the two different luciferases Nanoluc shows a superior signal overall and an improved signal-over-noise ratio. Hence, we selected the Nanoluc luciferase as our reporter system for our Universal test construct for all our future experiments. | Therefore, we went back to the design phase and decided to further look into luminescence-based reporters. The next experiment we performed was the comparison of the Firefly with the Nanoluc luciferase. Generally, it can be observed that the background of luminescence is very low in the control, compared to fluorescence measurements (Figure 2). Additionally, high luminescence levels can be observed, which allows the detection of successful expression from our chloroplast cell-free extracts. When comparing the two different luciferases Nanoluc shows a superior signal overall and an improved signal-over-noise ratio. Hence, we selected the Nanoluc luciferase as our reporter system for our Universal test construct for all our future experiments. | ||
| Line 69: | Line 70: | ||
</figure></center></html> | </figure></center></html> | ||
| + | <h3>Proof of Concept</h3> | ||
<p> | <p> | ||
This first version of our Universal test construct allowed us to develop, prototype and troubleshoot our first cell-free systems of tobacco and spinach (Figure 3). But we did not want to stop there and went though more rounds of the design-build-test-learn cycle and aimed to optimize this Universal test construct further, with the goal in mind to have a construct at hand which can even work for cell-free systems of non-model plant species. | This first version of our Universal test construct allowed us to develop, prototype and troubleshoot our first cell-free systems of tobacco and spinach (Figure 3). But we did not want to stop there and went though more rounds of the design-build-test-learn cycle and aimed to optimize this Universal test construct further, with the goal in mind to have a construct at hand which can even work for cell-free systems of non-model plant species. | ||
| Line 80: | Line 82: | ||
</figure></center></html> | </figure></center></html> | ||
| + | <h3>Codon Optimization</h3> | ||
<p> | <p> | ||
As we have now decided for the Nanoluc luciferase as a reporter we wanted to test if different codon optimizations would have an effect on the expression level in our system. Surprisingly, we found that our Nanoluc part, which is optimized for the chloroplast of Chlamydomonas reinhardtii, shows higher expression in a tobacco cell-free system, than a Nanoluc part specifically optimized for the chloroplast of Nicotiana tabacum (Figure 4). There are many possible explanations for that, one example would be the mRNA structure at the beginning of the coding region of the optimized Nanoluc parts. Due to the fact that we were just interested in the optimization of our construct for high expression, we chose the version, which is optimized for Chlamydomonas reinhardtii, for our next version of our Universal test construct. | As we have now decided for the Nanoluc luciferase as a reporter we wanted to test if different codon optimizations would have an effect on the expression level in our system. Surprisingly, we found that our Nanoluc part, which is optimized for the chloroplast of Chlamydomonas reinhardtii, shows higher expression in a tobacco cell-free system, than a Nanoluc part specifically optimized for the chloroplast of Nicotiana tabacum (Figure 4). There are many possible explanations for that, one example would be the mRNA structure at the beginning of the coding region of the optimized Nanoluc parts. Due to the fact that we were just interested in the optimization of our construct for high expression, we chose the version, which is optimized for Chlamydomonas reinhardtii, for our next version of our Universal test construct. | ||
| Line 91: | Line 94: | ||
</figure></center></html> | </figure></center></html> | ||
| − | + | <h3>Regulatory Elements</h3> | |
| + | <h4>5'UTR</h4> | ||
<p> | <p> | ||
The next element we aimed to optimize was the selection of the 5’UTR. For this we looked into literature again and compared two different 5’UTRs, which have been reported to enable high expression in vivo in the chloroplast of tobacco [reference]. When comparing these two 5’UTRs it can be observed that the 5’UTR of gene10, which originates from the E. coli T7 phage, shows far higher expression than the endogenous 5’UTR of rbcL (the large subunit of the RuBisCo). With that knowledge in mind we designed the next version of the Universal test construct with the aforementioned phage part. | The next element we aimed to optimize was the selection of the 5’UTR. For this we looked into literature again and compared two different 5’UTRs, which have been reported to enable high expression in vivo in the chloroplast of tobacco [reference]. When comparing these two 5’UTRs it can be observed that the 5’UTR of gene10, which originates from the E. coli T7 phage, shows far higher expression than the endogenous 5’UTR of rbcL (the large subunit of the RuBisCo). With that knowledge in mind we designed the next version of the Universal test construct with the aforementioned phage part. | ||
| Line 100: | Line 104: | ||
<figure> | <figure> | ||
<img src='https://2021.igem.org/wiki/images/0/0d/T--Marburg--Optimization_5%27UTR_UTC.png' width='700px' alt='5'UTR_Optimization' /> | <img src='https://2021.igem.org/wiki/images/0/0d/T--Marburg--Optimization_5%27UTR_UTC.png' width='700px' alt='5'UTR_Optimization' /> | ||
| − | <figcaption>test</figcaption> | + | <figcaption><b>Figure 5: Comparing different 5'UTR of the Universal test construct</b><br> |
| + | Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the <a href="https://parts.igem.org/Part:BBa_K3758304">Universal test construct 4.0</a> and the <a href="https://parts.igem.org/Part:BBa_K3758305">Universal test construct 5.0</a> including the gene10 5'UTR region of the T7 phage</figcaption> | ||
</figure></center></html> | </figure></center></html> | ||
| − | + | <h4>3'UTR</h4> | |
<p> | <p> | ||
The last part of the construct we wanted to optimize was the 3’UTR. For this we even went through two rounds of engineering and optimization. In the first experiment we compared the commonly used 3’UTR rrnB, which originates from E.coli with the psbA 3’UTR (photosystem II D1 protein) of Nicotiana tabacum. In this experiment it can be observed that our construct can already be significantly improved by exchanging the rrnB 3’UTR to the psbA 3’UTR. | The last part of the construct we wanted to optimize was the 3’UTR. For this we even went through two rounds of engineering and optimization. In the first experiment we compared the commonly used 3’UTR rrnB, which originates from E.coli with the psbA 3’UTR (photosystem II D1 protein) of Nicotiana tabacum. In this experiment it can be observed that our construct can already be significantly improved by exchanging the rrnB 3’UTR to the psbA 3’UTR. | ||
| Line 115: | Line 120: | ||
<figure> | <figure> | ||
<img src='https://2021.igem.org/wiki/images/5/5b/T--Marburg--Optimization_3%27UTR_UTC.png' width='700px' alt='3'UTR_Optimization' /> | <img src='https://2021.igem.org/wiki/images/5/5b/T--Marburg--Optimization_3%27UTR_UTC.png' width='700px' alt='3'UTR_Optimization' /> | ||
| − | <figcaption>test</figcaption> | + | <figcaption><b>Figure 6: Comparing different 3'UTR of the Universal test construct</b><br> |
| + | Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the <a href="https://parts.igem.org/Part:BBa_K3758305">Universal test construct 5.0</a> and the <a href="https://parts.igem.org/Part:BBa_K3758306">Universal test construct 6.0</a> including the 3'UTR region of psbA of <i>Nicotiana tabacum</i></figcaption> | ||
</figure></center></html> | </figure></center></html> | ||
| + | <h4>Viral 3'UTR</h4> | ||
<p> | <p> | ||
At that point we initially thought we would have the best possible Universal test construct for future screening of non-model cell-free extracts. But when we did the extensive part characterization of different 5’ and 3’ UTR (read more here), we were able to identify another even better 3’UTR (Figure 7), which again comes from a viral background, but in this case from the tobacco mosaic virus. We were very surprised by the results as there was no experimental evidence in literature that this part would be functional at all. This again highlights the big advantage of our high-throughput part characterization approach, utilizing our chloroplast cell-free system. | At that point we initially thought we would have the best possible Universal test construct for future screening of non-model cell-free extracts. But when we did the extensive part characterization of different 5’ and 3’ UTR (read more here), we were able to identify another even better 3’UTR (Figure 7), which again comes from a viral background, but in this case from the tobacco mosaic virus. We were very surprised by the results as there was no experimental evidence in literature that this part would be functional at all. This again highlights the big advantage of our high-throughput part characterization approach, utilizing our chloroplast cell-free system. | ||
| Line 126: | Line 133: | ||
<figure> | <figure> | ||
<img src='https://2021.igem.org/wiki/images/6/6c/T--Marburg--Viral_3%27UTR_UTC.png' width='900px' alt='Reiteration_3'UTR_Optimization' /> | <img src='https://2021.igem.org/wiki/images/6/6c/T--Marburg--Viral_3%27UTR_UTC.png' width='900px' alt='Reiteration_3'UTR_Optimization' /> | ||
| − | <figcaption>test</figcaption> | + | <figcaption><b>Figure 7: Comparing different 3’UTRs</b><br> |
| + | Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. These data were extracted from the larger data set of our large scale part characterisation experiment and the other parts were left out to present the data more clearly. This measurement was performed using the <a href="https://parts.igem.org/Part:BBa_K3758305">Universal test construct 5.0</a> and the <a href="https://parts.igem.org/Part:BBa_K3758301">Universal test construct 7.0</a> including the 3'UTR region of the tobacco mosaic virus</figcaption> | ||
</figure></center></html> | </figure></center></html> | ||
| + | <h3>English Oak Cell-Free Extract</h3> | ||
<p> | <p> | ||
We were super excited about our results, that after all that optimization of our Universal test construct 7.0, we were able to develop a fully functional chloroplast extract of the english oak tree and were even able to test other parts in this system. This would not have been possible without all of these iterations of the design-build-test-learn cycle, as even our version 5.0 of the test device showed far less expression. | We were super excited about our results, that after all that optimization of our Universal test construct 7.0, we were able to develop a fully functional chloroplast extract of the english oak tree and were even able to test other parts in this system. This would not have been possible without all of these iterations of the design-build-test-learn cycle, as even our version 5.0 of the test device showed far less expression. | ||
| Line 137: | Line 146: | ||
<figure> | <figure> | ||
<img src='https://2021.igem.org/wiki/images/2/21/T--Marburg--Oak_Proof_of_Concept_UTC.png' width='700px' alt='POC_Oak' /> | <img src='https://2021.igem.org/wiki/images/2/21/T--Marburg--Oak_Proof_of_Concept_UTC.png' width='700px' alt='POC_Oak' /> | ||
| − | <figcaption> | + | <figcaption><b>Figure 8: Successful Expression using the Universal Test Construct 7.0 in a Cell-Free Extract of <i>Quercus robur</i></b><br> |
| + | Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 160µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurment has been performed using our best composite part <a href="https://parts.igem.org/Part:BBa_K3758301">Universal Test Construct 7.0</a> and comparing it to the version <a href="https://parts.igem.org/Part:BBa_K3758305">Universal Test Construct 5.0</a></figcaption> | ||
</figure></center></html> | </figure></center></html> | ||
| + | |||
| + | <h3>Conclusion</h3> | ||
| + | <p> | ||
| + | All in all, we hope that future teams could use our knowledge, but especially our latest version of our Universal test construct 7.0 to develop complete new project ideas, involving any plant chassis they could think of. By using our best new composite part, future teams can already skip the “chicken or egg” problem, which we described at the beginning and start troubleshooting their extract preparation without worrying about designing-building and testing their own test device in the first place. | ||
| + | </p> | ||
Latest revision as of 16:49, 18 October 2021
T7 Universal Test Construct 7.0
Contents
Part Description
We proudly present our best new composite part:
The Universal T7 Test Construct 7.0. After seven rounds of the design-build-test-learn cycle, we have identified the most efficient combination of regulatory parts and reporter genes for the expression in chloroplast cell-free extracts. This composite part allowed us to develop the first fully functional cell-free extract of english oak chloroplasts, which would not have been possible without our engineering iterations. We hope that future teams could use our Universal test construct 7.0 to develop complete new project ideas, involving any plant chassis they could think of and directly start troubleshooting their extract preparation without worrying about the optimal DNA construct design in the first place.
When we started our project we had to face a typical “chicken or egg” problem: How would we design our first test construct in order to verify that we have a working chloroplast cell-free extract without having a working extract where we can test if our test construct design works. For this we tried to use one of engineering principles, which was proposed in the early days of synthetic biology: Decoupling. Decoupling is the concept of dividing a complex problem into many simpler problems that can be worked on separately[18]. Although this principle is not 100% translatable to some biological engineering problems, as many things are interconnected and influence each other, it still helped us to overcome the “chicken or egg” barrier. The following basic parts would are required in order to build and test a functional transcriptional unit for our chloroplast extracts:
- Promoter
- 5’untranslated region (UTR)
- Reporter gene/codon optimization of the reporter
- 3‘untranslated region (UTR)
For the promoter we decided on using the T7 promoter for our universal test construct, which requires the addition of the T7 polymerase to the cell- free reaction. The usage of the T7 transcription system allowed us to reduce the number of possible problems, due to the fact that we have more control over it as we can just titrate the amount of T7 polymerase for our cell-free reaction. For the other positions, we went through multiple iterations of the Design-Build-Test-Learn Cycle and optimized our test construct for highest possible expression. The optimization can be found below under the section Results.
Characterization & Measurement
Batch effect
A major difficulty when working with cell free technology is the reproducibility of reliable data generation. While one batch of cell free extract generation might be perfectly suited for efficient protein production, other batches might not perform that well. Because our chloroplast extracts were prepared from leaves, each preparation could have contained distinct compositions of differentiated leaf cells (for example: mesophyll, palisade parenchyma and bundle sheath) from plants that experienced microclimatic variations. This in turns causes the individual difference in expression strength, making it impossible to quantitatively compare data across measurements.
Results
Choosing the Reporter System
Fluorescence
The next important design decision was the selection of the reporter system we would use. Therefore, we looked into different fluorescent proteins, such as mScarlet and sfGFP, but also into luciferase-based reporters, like the Firefly and Nanoluc luciferase. After our first measurements, it was clear that fluorescent reporters are not suitable for our cell-free measurements in our chloroplast systems, as the signal can not be distinguished from the noise (Figure 1). On the one hand, this can be explained due to the autofluorescence of the remaining chlorophyll molecules, which increase the fluorescence background levels of the extract control. And on the other hand the expression level of our cell-free reaction seems to be too low at least for the beginning, when we were still at the troubleshooting phase of our extract preparation.
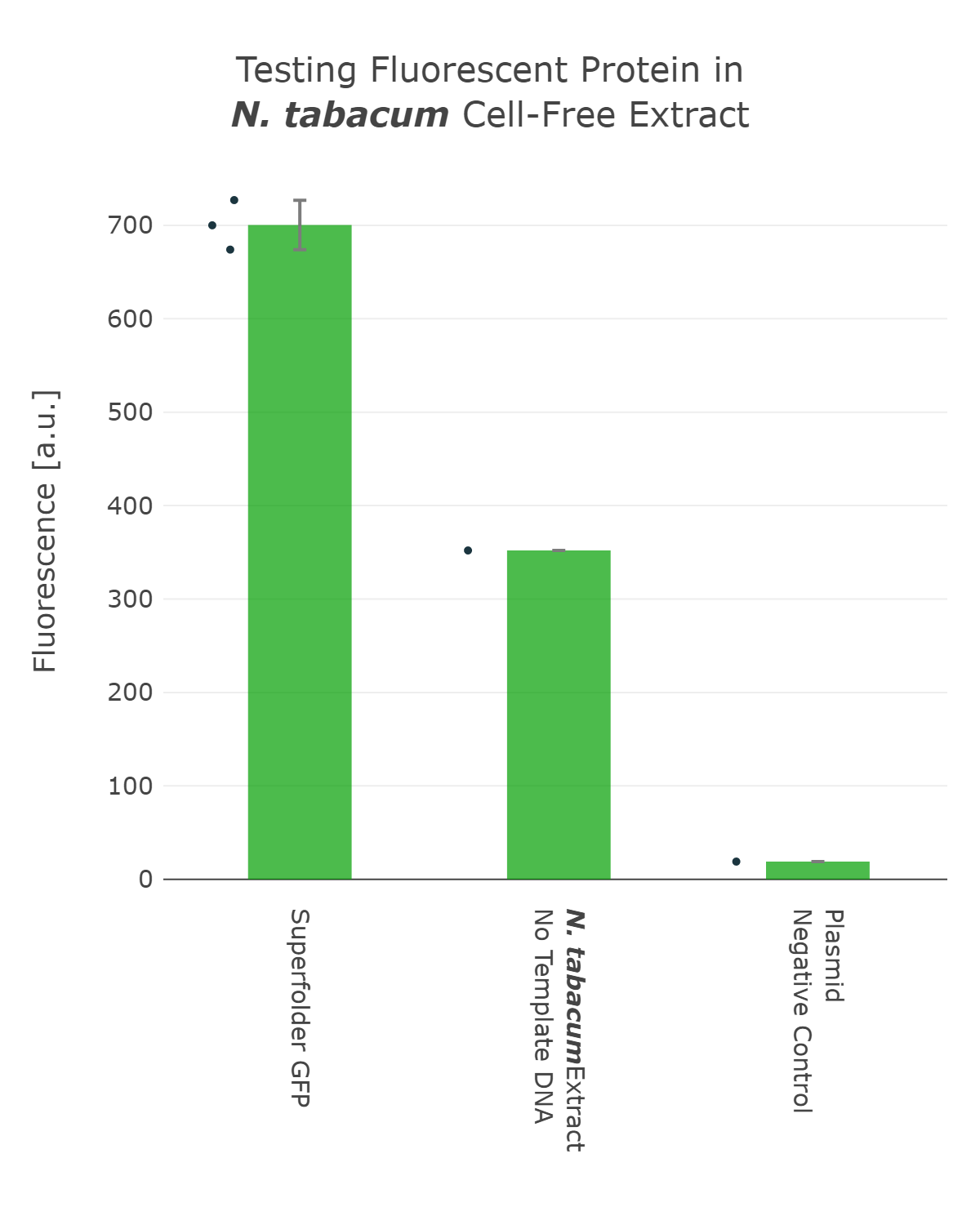
Fluorescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively. This measurement was performed using the first version of our Universal test construct including sfGFP
Luminescence
Therefore, we went back to the design phase and decided to further look into luminescence-based reporters. The next experiment we performed was the comparison of the Firefly with the Nanoluc luciferase. Generally, it can be observed that the background of luminescence is very low in the control, compared to fluorescence measurements (Figure 2). Additionally, high luminescence levels can be observed, which allows the detection of successful expression from our chloroplast cell-free extracts. When comparing the two different luciferases Nanoluc shows a superior signal overall and an improved signal-over-noise ratio. Hence, we selected the Nanoluc luciferase as our reporter system for our Universal test construct for all our future experiments.
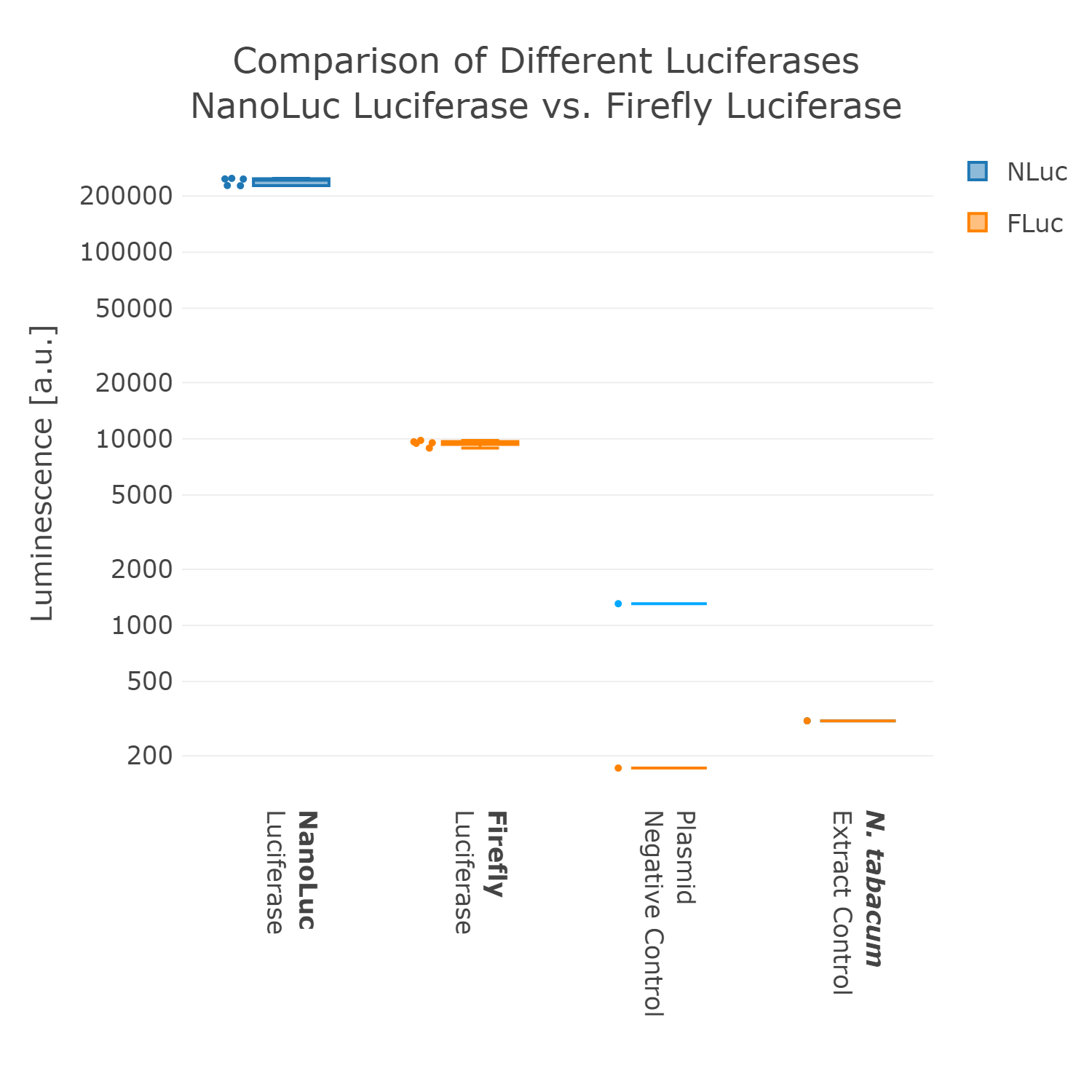
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up in a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression.This measurement was performed using the Universal test construct 2.0 including the Firefly luciferase and Universal test construct 3.0 including the NanoLuc luciferase
Proof of Concept
This first version of our Universal test construct allowed us to develop, prototype and troubleshoot our first cell-free systems of tobacco and spinach (Figure 3). But we did not want to stop there and went though more rounds of the design-build-test-learn cycle and aimed to optimize this Universal test construct further, with the goal in mind to have a construct at hand which can even work for cell-free systems of non-model plant species.
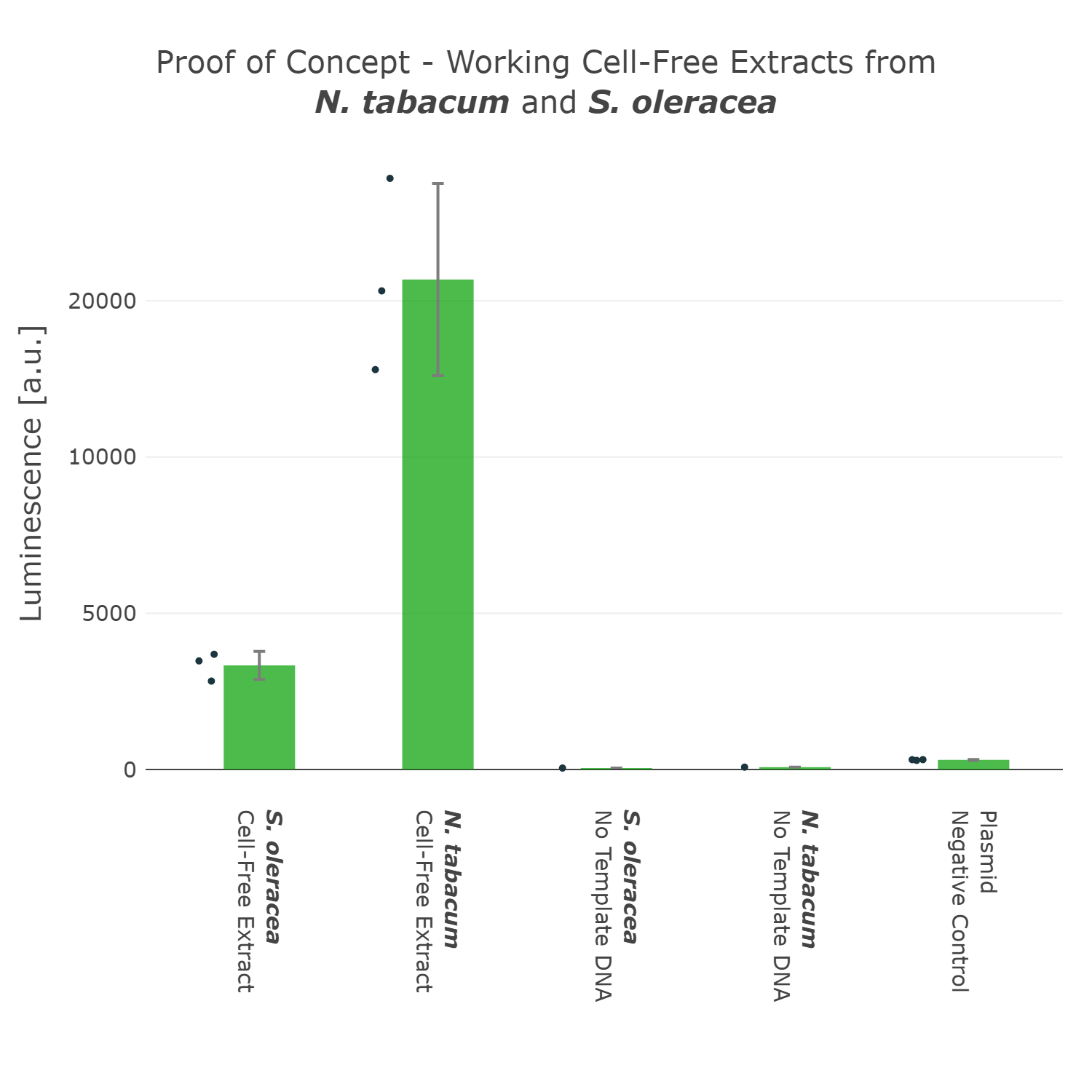
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up in a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the Universal test construct 3.0 including the NanoLuc luciferase
Codon Optimization
As we have now decided for the Nanoluc luciferase as a reporter we wanted to test if different codon optimizations would have an effect on the expression level in our system. Surprisingly, we found that our Nanoluc part, which is optimized for the chloroplast of Chlamydomonas reinhardtii, shows higher expression in a tobacco cell-free system, than a Nanoluc part specifically optimized for the chloroplast of Nicotiana tabacum (Figure 4). There are many possible explanations for that, one example would be the mRNA structure at the beginning of the coding region of the optimized Nanoluc parts. Due to the fact that we were just interested in the optimization of our construct for high expression, we chose the version, which is optimized for Chlamydomonas reinhardtii, for our next version of our Universal test construct.
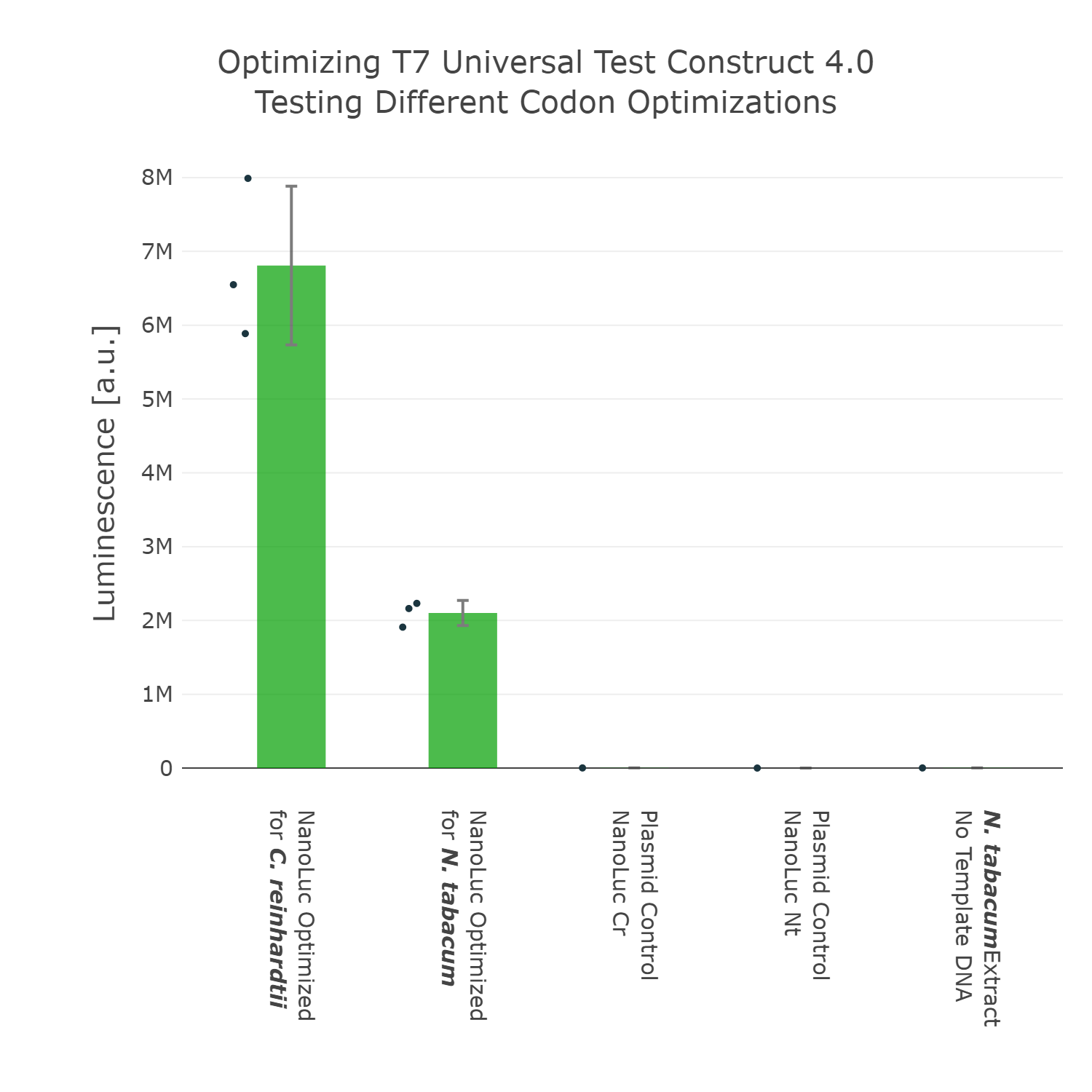
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the Universal test construct 3.0 including the NanoLuc luciferase codon optimized for the chloroplast of Nicotiana tabacum and the Universal test construct 4.0 including the NanoLuc luciferase codon optimized for the chloroplast of Chlamydomonas reinhardtii
Regulatory Elements
5'UTR
The next element we aimed to optimize was the selection of the 5’UTR. For this we looked into literature again and compared two different 5’UTRs, which have been reported to enable high expression in vivo in the chloroplast of tobacco [reference]. When comparing these two 5’UTRs it can be observed that the 5’UTR of gene10, which originates from the E. coli T7 phage, shows far higher expression than the endogenous 5’UTR of rbcL (the large subunit of the RuBisCo). With that knowledge in mind we designed the next version of the Universal test construct with the aforementioned phage part.
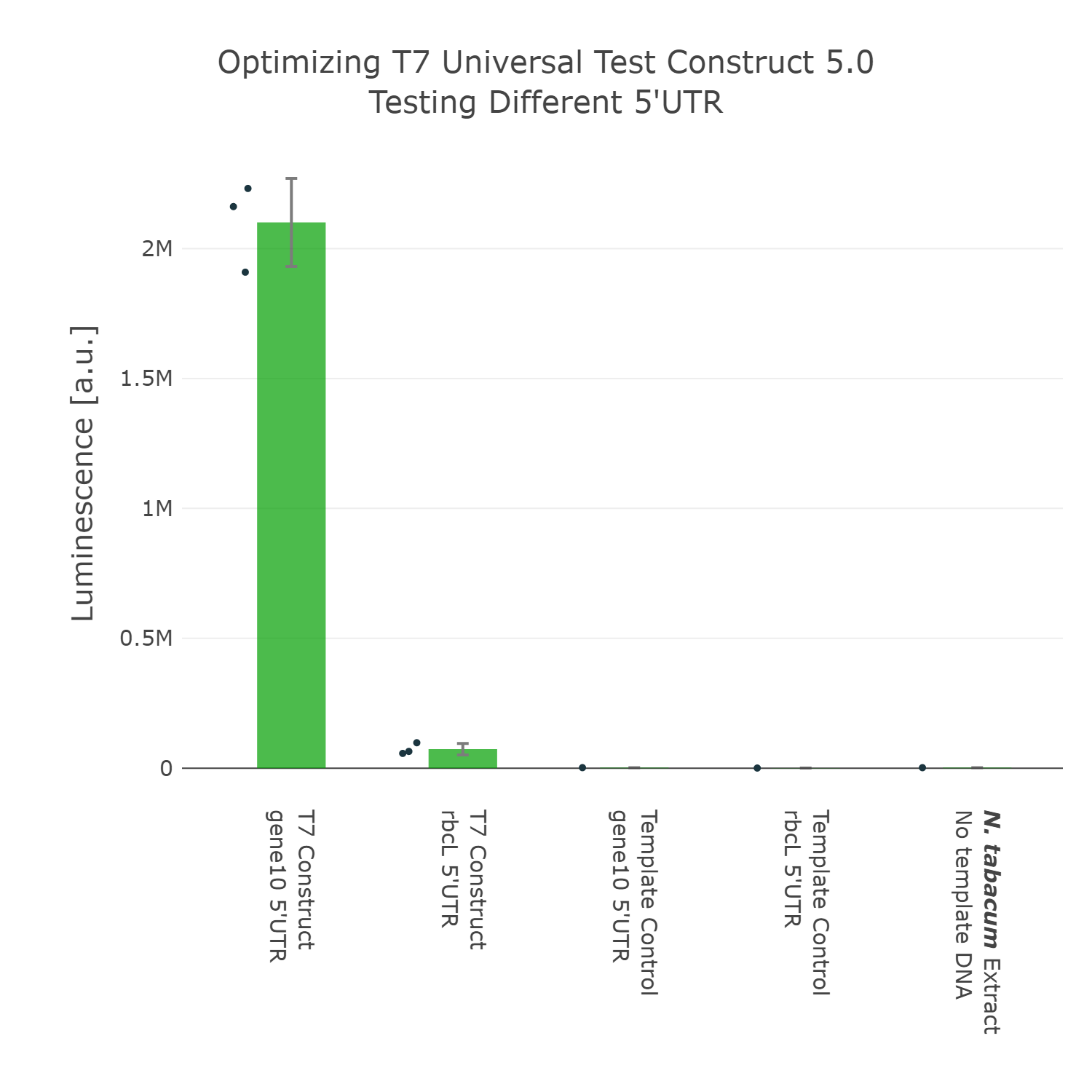
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the Universal test construct 4.0 and the Universal test construct 5.0 including the gene10 5'UTR region of the T7 phage
3'UTR
The last part of the construct we wanted to optimize was the 3’UTR. For this we even went through two rounds of engineering and optimization. In the first experiment we compared the commonly used 3’UTR rrnB, which originates from E.coli with the psbA 3’UTR (photosystem II D1 protein) of Nicotiana tabacum. In this experiment it can be observed that our construct can already be significantly improved by exchanging the rrnB 3’UTR to the psbA 3’UTR.
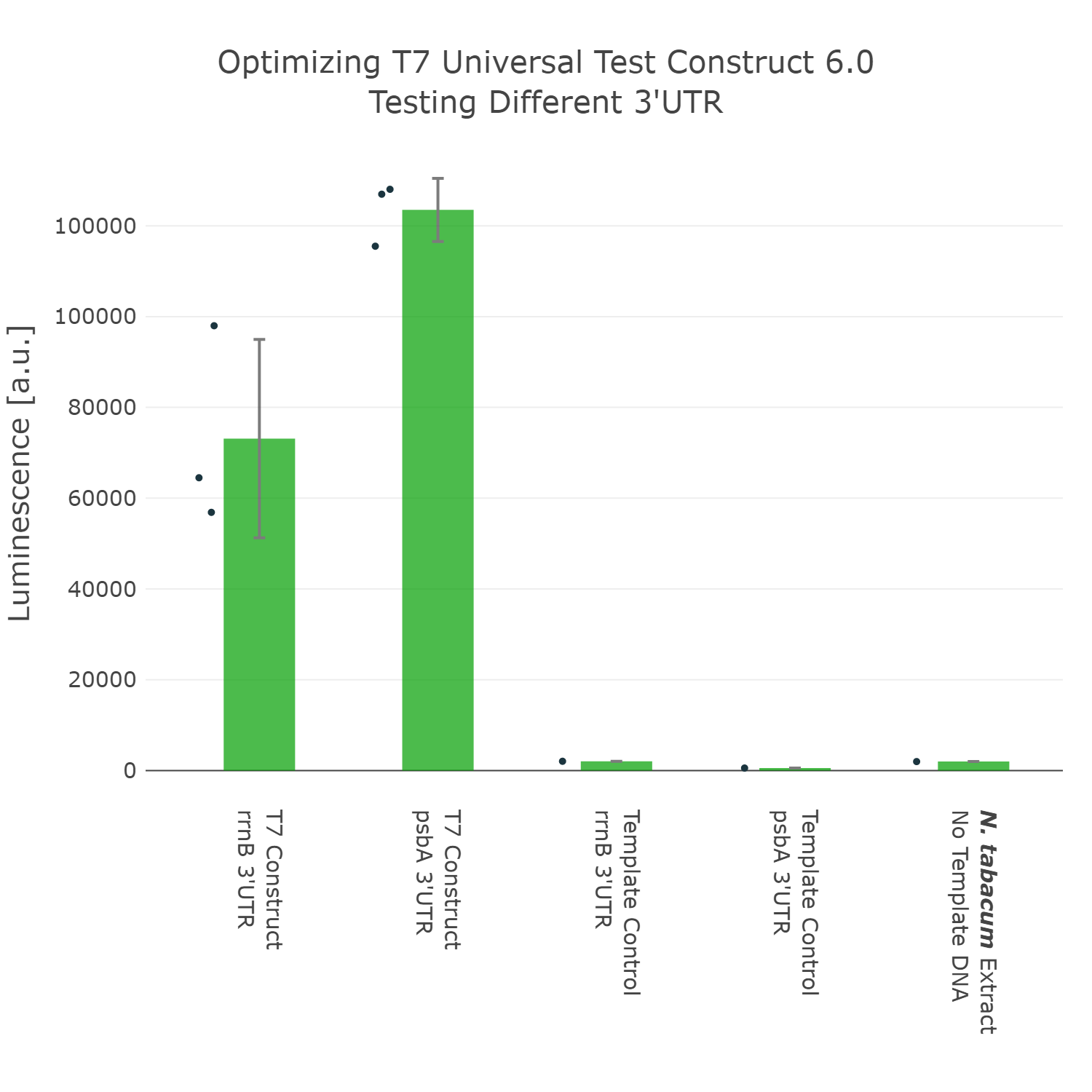
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurement was performed using the Universal test construct 5.0 and the Universal test construct 6.0 including the 3'UTR region of psbA of Nicotiana tabacum
Viral 3'UTR
At that point we initially thought we would have the best possible Universal test construct for future screening of non-model cell-free extracts. But when we did the extensive part characterization of different 5’ and 3’ UTR (read more here), we were able to identify another even better 3’UTR (Figure 7), which again comes from a viral background, but in this case from the tobacco mosaic virus. We were very surprised by the results as there was no experimental evidence in literature that this part would be functional at all. This again highlights the big advantage of our high-throughput part characterization approach, utilizing our chloroplast cell-free system.
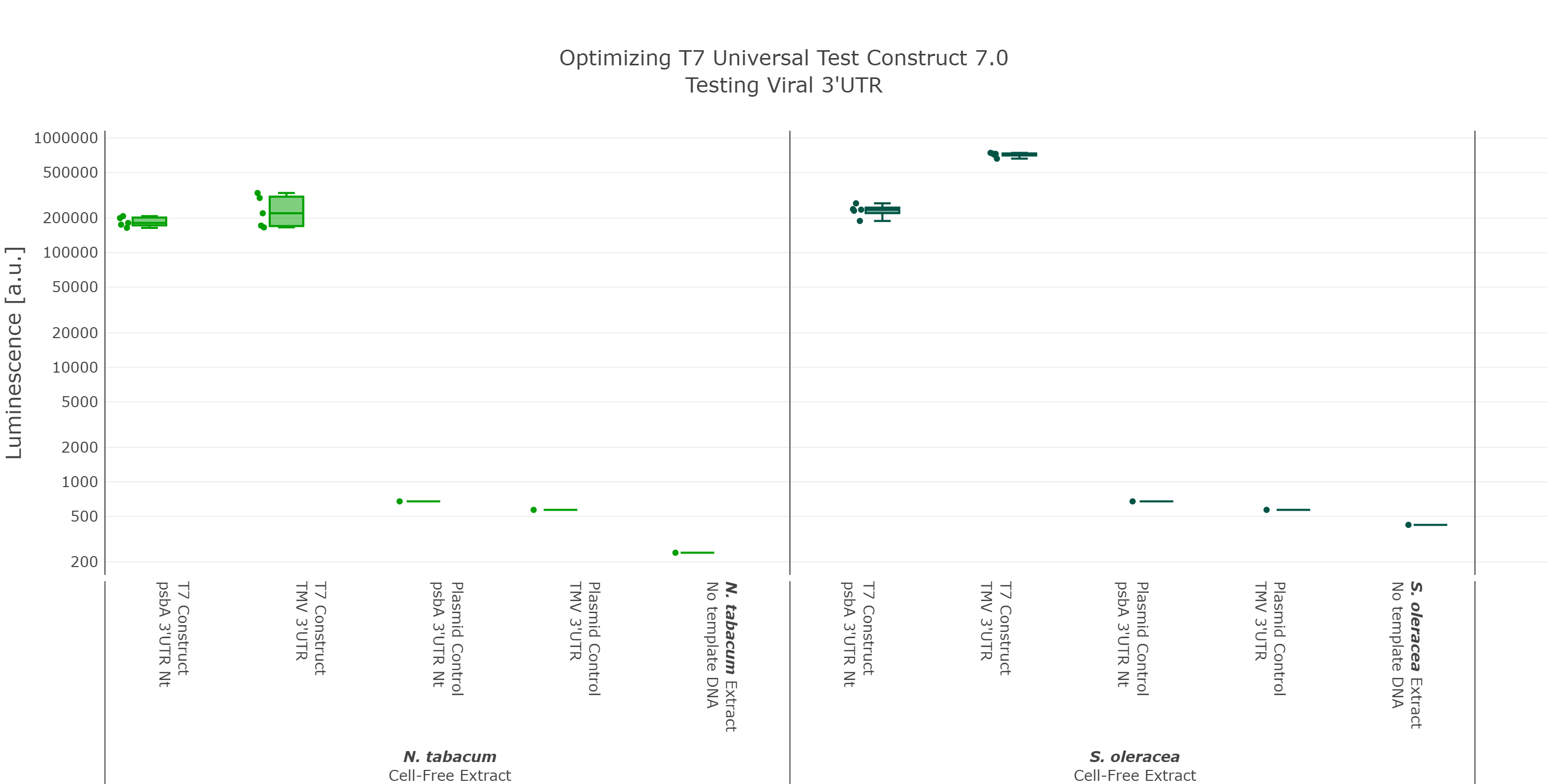
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 10µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. These data were extracted from the larger data set of our large scale part characterisation experiment and the other parts were left out to present the data more clearly. This measurement was performed using the Universal test construct 5.0 and the Universal test construct 7.0 including the 3'UTR region of the tobacco mosaic virus
English Oak Cell-Free Extract
We were super excited about our results, that after all that optimization of our Universal test construct 7.0, we were able to develop a fully functional chloroplast extract of the english oak tree and were even able to test other parts in this system. This would not have been possible without all of these iterations of the design-build-test-learn cycle, as even our version 5.0 of the test device showed far less expression.
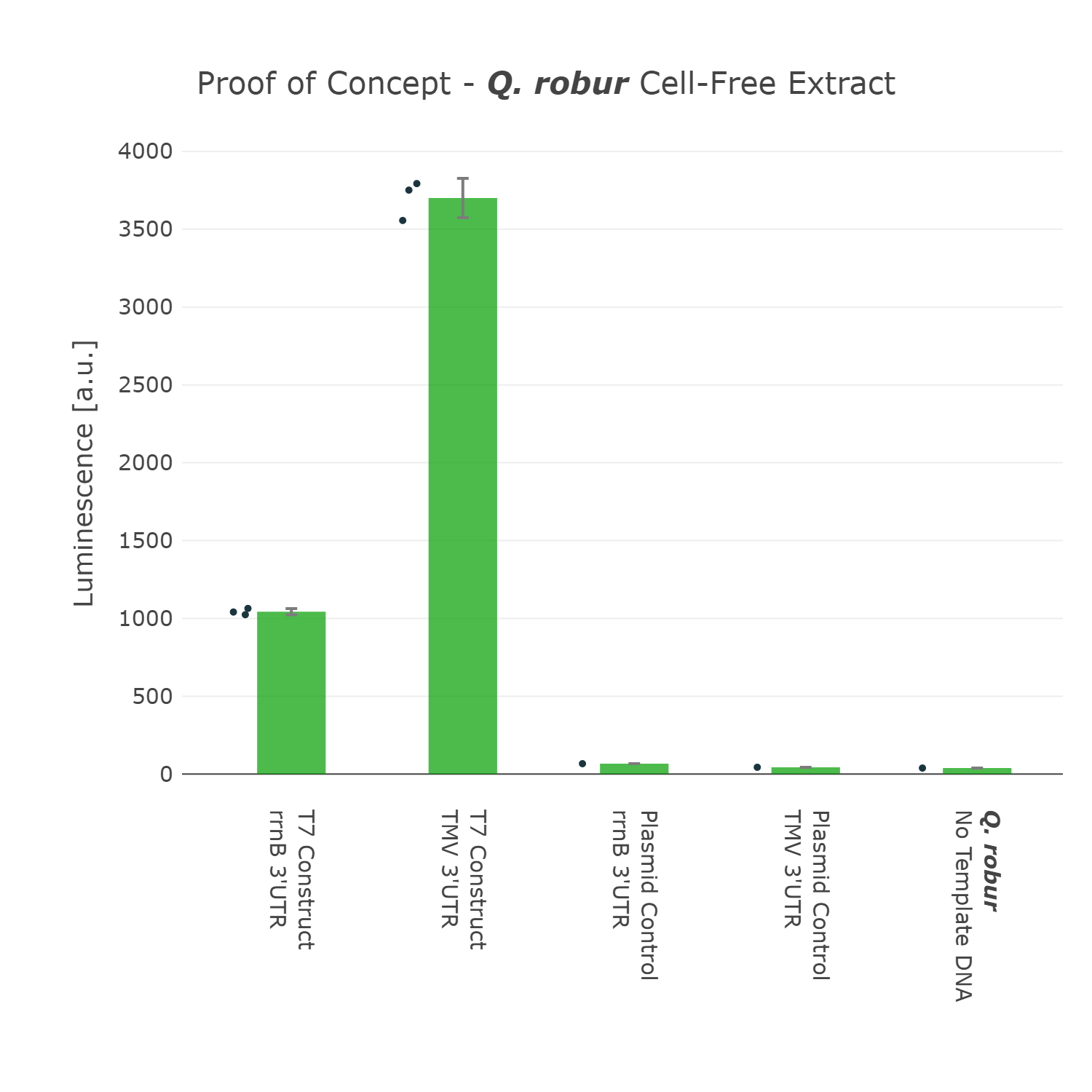
Luminescence values are given as arbitrary units and the data is presented on a logarithmic scale. The reaction was set up with a total volume of 160µl. Negative controls using only the plasmid DNA or the crude chloroplast extracts have been included respectively in order to verify the expression. This measurment has been performed using our best composite part Universal Test Construct 7.0 and comparing it to the version Universal Test Construct 5.0
Conclusion
All in all, we hope that future teams could use our knowledge, but especially our latest version of our Universal test construct 7.0 to develop complete new project ideas, involving any plant chassis they could think of. By using our best new composite part, future teams can already skip the “chicken or egg” problem, which we described at the beginning and start troubleshooting their extract preparation without worrying about designing-building and testing their own test device in the first place.
Marburg collection 3.0
We proudly present the third expansion of the Marburg collection [15]. The Marburg collection is a Golden Gate based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. The collection overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units.
The original [http://2018.igem.org/Team:Marburg/Part_Collection Marburg Collection] contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. The toolbox was constructed as a foundation for future iGEM teams to empower accelerated progression in their ambitious projects.
Our collection includes genetic parts suitable for use in the chloroplast. Among parts from the chloroplast of Nicotiana tabacum, we built parts from the chloroplast of Spinacia oleracea, Oryza sativa,Triticum aestivum and Quercus robur. With our contribution, we aim to accelerate research in the field of plastid engineering.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 59
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 4
- 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 59
- 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 59
- 1000COMPATIBLE WITH RFC[1000]
References
[1] Aboul-Maaty, N. A.-F., & Oraby, H. A.-S. (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bulletin of the National Research Centre, 43(1). https://doi.org/10.1186/s42269-019-0066-1
[2] Fuentes, P., Zhou, F., Erban, A., Karcher, D., Kopka, J., & Bock, R. (2016). A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. ELife, 5. https://doi.org/10.7554/elife.13664
[3] Filée, J., & Forterre, P. (2005). Viral proteins functioning in organelles: a cryptic origin? Trends in Microbiology, 13(11), 510–513. https://doi.org/10.1016/j.tim.2005.08.012
[4] Liere, K., & Börner, T. (2007). Transcription and transcriptional regulation in plastids. In Cell and Molecular Biology of Plastids (pp. 121–174). Springer Berlin Heidelberg. https://doi.org/10.1007/4735_2007_0232
[5] Xie, G., & Allison, L. (2002). Sequences upstream of the YRTA core region are essential for transcription of the tobacco atpB NEP promoter in chloroplasts in vivo. Current Genetics, 41(3), 176–182. https://doi.org/10.1007/s00294-002-0293-z
[6] Kuroda, H., & Maliga, P. (2002). Overexpression of the clpP 5′-Untranslated Region in a Chimeric Context Causes a Mutant Phenotype, Suggesting Competition for a clpP-Specific RNA Maturation Factor in Tobacco Chloroplasts. Plant Physiology, 129(4), 1600–1606. https://doi.org/10.1104/pp.004986
[7] Klinkert, B. (2006). Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Research, 34(1), 386–394. https://doi.org/10.1093/nar/gkj433
[8] Zhelyazkova, P., Hammani, K., Rojas, M., Voelker, R., Vargas-Suárez, M., Börner, T., & Barkan, A. (2011). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Research, 40(7), 3092–3105. https://doi.org/10.1093/nar/gkr1137
[9] Zoschke, R., Kroeger, T., Belcher, S., Schöttler, M. A., Barkan, A., & Schmitz-Linneweber, C. (2012). The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplastatpB/EmRNA. The Plant Journal, 72(4), 547–558. https://doi.org/10.1111/j.1365-313x.2012.05081.x
[10] Zhang, L., Zhou, W., Che, L., Rochaix, J.-D., Lu, C., Li, W., & Peng, L. (2019). PPR Protein BFA2 Is Essential for the Accumulation of the atpH/F Transcript in Chloroplasts. Frontiers in Plant Science, 10. https://doi.org/10.3389/fpls.2019.00446
[11] Tangphatsornruang, S., Birch-Machin, I., Newell, C. A., & Gray, J. C. (2010). The effect of different 3′ untranslated regions on the accumulation and stability of transcripts of a gfp transgene in chloroplasts of transplastomic tobacco. Plant Molecular Biology, 76(3–5), 385–396. https://doi.org/10.1007/s11103-010-9689-1
[12] Eberhard, S., Drapier, D., & Wollman, F.-A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. The Plant Journal, 31(2), 149–160. https://doi.org/10.1046/j.1365-313x.2002.01340.x
[13] Pfalz, J., Bayraktar, O. A., Prikryl, J., & Barkan, A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. The EMBO Journal, 28(14), 2042–2052. https://doi.org/10.1038/emboj.2009.121
[14] Schaumberg, K. A., Antunes, M. S., Kassaw, T. K., Xu, W., Zalewski, C. S., Medford, J. I., & Prasad, A. (2015). Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nature Methods, 13(1), 94–100. https://doi.org/10.1038/nmeth.3659
[15] Stukenberg, D., Hensel, T., Hoff, J., Daniel, B., Inckemann, R., Tedeschi, J. N., Nousch, F., & Fritz, G. (2021). The Marburg Collection: A Golden Gate DNA Assembly Framework for Synthetic Biology Applications in Vibrio natriegens. ACS Synthetic Biology, 10(8), 1904–1919. https://doi.org/10.1021/acssynbio.1c00126
[16] Suzuki, J. Y., Sriraman, P., Svab, Z., & Maliga, P. (2003). Unique Architecture of the Plastid Ribosomal RNA Operon Promoter Recognized by the Multisubunit RNA Polymerase in Tobacco and Other Higher Plants. The Plant Cell, 15(1), 195–205. https://doi.org/10.1105/tpc.007914
[17] Occhialini, A., Piatek, A. A., Pfotenhauer, A. C., Frazier, T. P., Stewart, C. N., Jr., & Lenaghan, S. C. (2019). MoChlo: A Versatile, Modular Cloning Toolbox for Chloroplast Biotechnology. Plant Physiology, 179(3), 943–957. https://doi.org/10.1104/pp.18.01220
[18]Endy, D. (2005). Foundations for engineering biology. In Nature (Vol. 438, Issue 7067, pp. 449–453). Springer Science and Business Media LLC. https://doi.org/10.1038/nature04342
