Difference between revisions of "Part:BBa K4090000"
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K4090000 short</partinfo> | <partinfo>BBa_K4090000 short</partinfo> | ||
| + | ==Introduction== | ||
| + | INP protein is a transmembrane protein which enables Gram-negative bacteria to promote nucleation of ice at relatively high temperatures. E.coli does not have the gene that encodes this kind of protein. However, research shows that the protein could be successully express on the plasma membrane of E.coli. The co-expression of the INP and other genes could bring the proteins that other genes encode to the cell surface. This biobrick contains the sequence of INP gene after the codon optimization base on the codon bias of E.coli. | ||
| − | + | <!-- --> | |
| − | + | <partinfo>BBa_K4090000 SequenceAndFeatures</partinfo> | |
<!-- Add more about the biology of this part here--> | <!-- Add more about the biology of this part here--> | ||
| − | == | + | ==Usage and Viability== |
| − | [[File:T-- | + | In order to prove the function of the INP, the experiment was performed, which co-expresses the INP gene with the silicatein gene. Silicatein is a kind of protein which can induce the precipitation of silica in favorable environment. |
| − | + | <br> | |
| − | [[File:T--SDSZ China--result 1-4parts.png| | + | [[File:T--SDSZ China--result 1-3parts.png|500px|thumb|center|Fig.1]] |
| + | <br><br> | ||
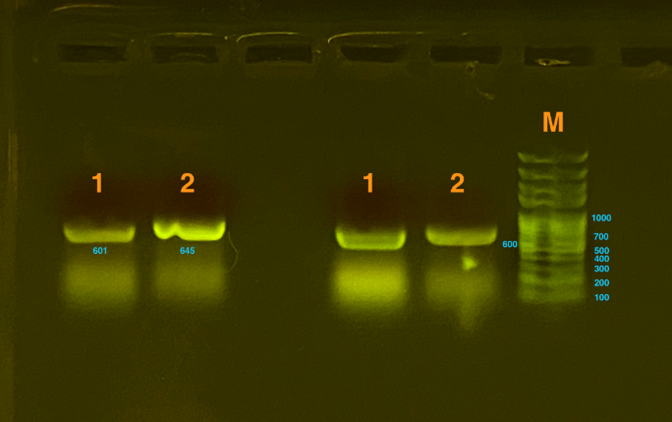
| + | Gibson Assembly was used to transform the gene. Then, the gel electrophoresis was used to test whether plasmids were constructed successfully. Since the gene that inserted is too long to detect as a whole, which is about 2000bp, our team decided to detect sequences of two connections (one connected the silicatein with the vector and the other connected the INP with silicatein) , both of which are approximately 600bp. From the graph below, we could see that the result was positive, proving that the plasmids are successfully constructed. | ||
| + | [[File:T--SDSZ China--result 1-4parts.png|500px|thumb|center|Fig.2]] | ||
Fig.1-2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome. | Fig.1-2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome. | ||
| − | [[File:T--SDSZ China--result 1-6parts.png| | + | <br><br> |
| + | After using the Gibson Assembly to transform the INP-silicatein gene in the E.coli and cultured them under appropriate environment, controlled experiment is performed. In order to know whether the silicatein was also expressed inside the cell, part of the E.coli were homogenized by ultrasonic comminution and added to the sample. The graph below showed that the experimental groups present higher OD values, and the group with homogenization has higher OD values than the group without homogenization. The result showed that INP could be used to bring the protein to the surface, though there is still a small amount of protein could not be expressed on the surface. | ||
| + | [[File:T--SDSZ China--result 1-6parts.png|550px|thumb|center|Fig.3]] | ||
Fig.1-3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since: | Fig.1-3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since: | ||
•Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids). | •Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids). | ||
•Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs). | •Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs). | ||
| − | + | <br><br> | |
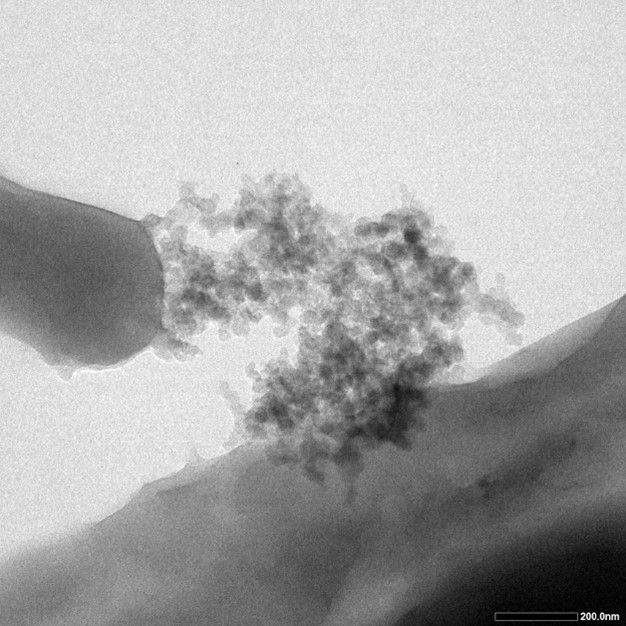
| − | + | TEM testing is also utilized in order to verify its function. Five drips of bacteria mixture of different concentration were added onto the five pieces of carbon film, and were put into the 80 degree Celsius dryer. After they are completely dry, TEM testing was carried out. The dark particles on the surface of cells, which are the silcia particles, further verified that the INP could bring the protein to the cell surface. | |
| − | [[File:T--SDSZ China--result 1- | + | [[File:T--SDSZ China--result 1-7parts.jpeg|300px|thumb|center|Fig.4]] |
| + | |||
| − | |||
| − | |||
==References== | ==References== | ||
| − | [1]ZHU T, PAULO C, MERROUN, M L, et al. Potential application of biomineralization by Synechococcus PCC8806 for concrete restoration[J]. Sedimentology, 2013,61(1):1-21<br> | + | [1] ZHU T, PAULO C, MERROUN, M L, et al. Potential application of biomineralization by Synechococcus PCC8806 for concrete restoration[J]. Sedimentology, 2013, 61(1): 1-21.<br> |
| − | [2]Lechner, Carolin C., and Christian FW Becker. "A sequence‐function analysis of the silica precipitating silaffin R5 peptide." Journal of Peptide Science 20.2 (2014): 152-158.<br> | + | [2] Lechner, Carolin C., and Christian FW Becker. "A sequence‐function analysis of the silica precipitating silaffin R5 peptide." Journal of Peptide Science 20.2 (2014): 152-158.<br> |
| − | [3]Poulsen, Nicole, and Nils Kroger. "Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana." Journal of Biological Chemistry 279.41 (2004): 42993-42999. | + | [3] Poulsen, Nicole, and Nils Kroger. "Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana." Journal of Biological Chemistry 279.41 (2004): 42993-42999. |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 19:12, 21 October 2021
Ice Nucleation Protein(INP)
Introduction
INP protein is a transmembrane protein which enables Gram-negative bacteria to promote nucleation of ice at relatively high temperatures. E.coli does not have the gene that encodes this kind of protein. However, research shows that the protein could be successully express on the plasma membrane of E.coli. The co-expression of the INP and other genes could bring the proteins that other genes encode to the cell surface. This biobrick contains the sequence of INP gene after the codon optimization base on the codon bias of E.coli.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 321
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 63
Illegal NgoMIV site found at 396
Illegal AgeI site found at 498 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 229
Usage and Viability
In order to prove the function of the INP, the experiment was performed, which co-expresses the INP gene with the silicatein gene. Silicatein is a kind of protein which can induce the precipitation of silica in favorable environment.
Gibson Assembly was used to transform the gene. Then, the gel electrophoresis was used to test whether plasmids were constructed successfully. Since the gene that inserted is too long to detect as a whole, which is about 2000bp, our team decided to detect sequences of two connections (one connected the silicatein with the vector and the other connected the INP with silicatein) , both of which are approximately 600bp. From the graph below, we could see that the result was positive, proving that the plasmids are successfully constructed.
Fig.1-2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome.
After using the Gibson Assembly to transform the INP-silicatein gene in the E.coli and cultured them under appropriate environment, controlled experiment is performed. In order to know whether the silicatein was also expressed inside the cell, part of the E.coli were homogenized by ultrasonic comminution and added to the sample. The graph below showed that the experimental groups present higher OD values, and the group with homogenization has higher OD values than the group without homogenization. The result showed that INP could be used to bring the protein to the surface, though there is still a small amount of protein could not be expressed on the surface.
Fig.1-3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since:
•Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids).
•Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs).
TEM testing is also utilized in order to verify its function. Five drips of bacteria mixture of different concentration were added onto the five pieces of carbon film, and were put into the 80 degree Celsius dryer. After they are completely dry, TEM testing was carried out. The dark particles on the surface of cells, which are the silcia particles, further verified that the INP could bring the protein to the cell surface.
References
[1] ZHU T, PAULO C, MERROUN, M L, et al. Potential application of biomineralization by Synechococcus PCC8806 for concrete restoration[J]. Sedimentology, 2013, 61(1): 1-21.
[2] Lechner, Carolin C., and Christian FW Becker. "A sequence‐function analysis of the silica precipitating silaffin R5 peptide." Journal of Peptide Science 20.2 (2014): 152-158.
[3] Poulsen, Nicole, and Nils Kroger. "Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana." Journal of Biological Chemistry 279.41 (2004): 42993-42999.