Difference between revisions of "Part:BBa K3962339"
| (3 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
https://2021.igem.org/Team:Leiden | https://2021.igem.org/Team:Leiden | ||
| − | We used this construct to calibrate the transcriptional activity of inducible promoter pBAD ([ | + | We used this construct to calibrate the transcriptional activity of inducible promoter pBAD ([https://parts.igem.org/Part:BBa_I0500 BBa_I0500]) by comparing its fluorescence intensity with pBAD::mCherry ([https://parts.igem.org/Part:BBa_K3962340 BBa_K3962340]) (Fig 1.). |
[[Image:T-Leiden-parts-Figuer1.png|500px]] | [[Image:T-Leiden-parts-Figuer1.png|500px]] | ||
Latest revision as of 11:55, 18 October 2021
J23100 promoter with mCherry for constant expression
The J23100 constitutive promoter is ligated to mCherry for the constant expression of fluorescent proteins. In our project, we used this construct as reference to calibrate the inducibility of L-arabinose to pBAD promoter activity.
2021 Team Leiden
https://2021.igem.org/Team:Leiden
We used this construct to calibrate the transcriptional activity of inducible promoter pBAD (BBa_I0500) by comparing its fluorescence intensity with pBAD::mCherry (BBa_K3962340) (Fig 1.).
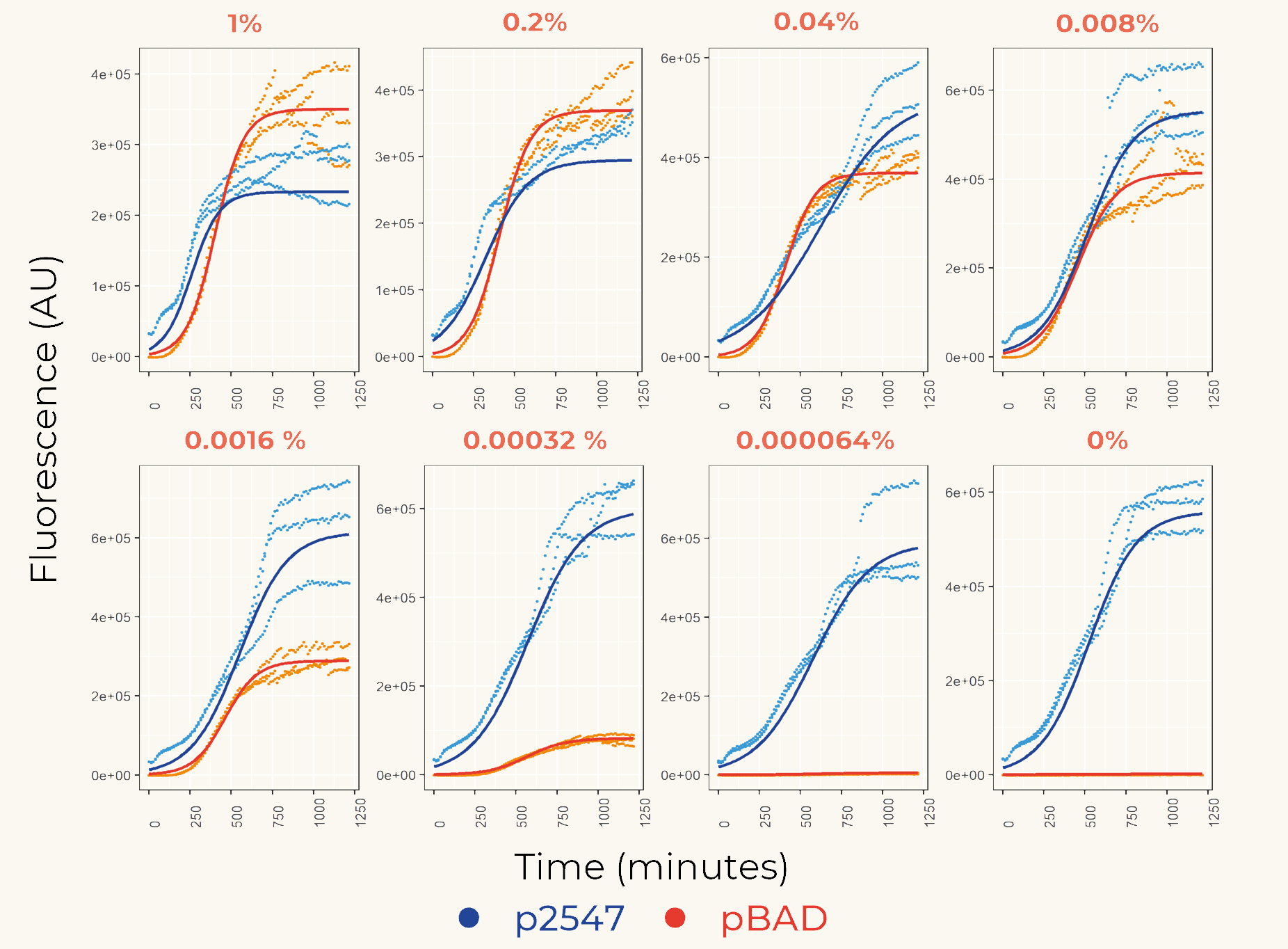
Figure 1. The fluorescence intensity of strains carrying our construct under different arabinose concentration. The measurement was carried out for 20 h. The x-axis shows the time of each measurement. The y-axis shows the fluorescence of mCherry in the arbitrary unit (AU). The blue data points represent the fluorescent intensity of p2547::mCherry strain, red data points represent the pBAD::mCherry strain. The arabinose concentration is shown at the top of each graph.
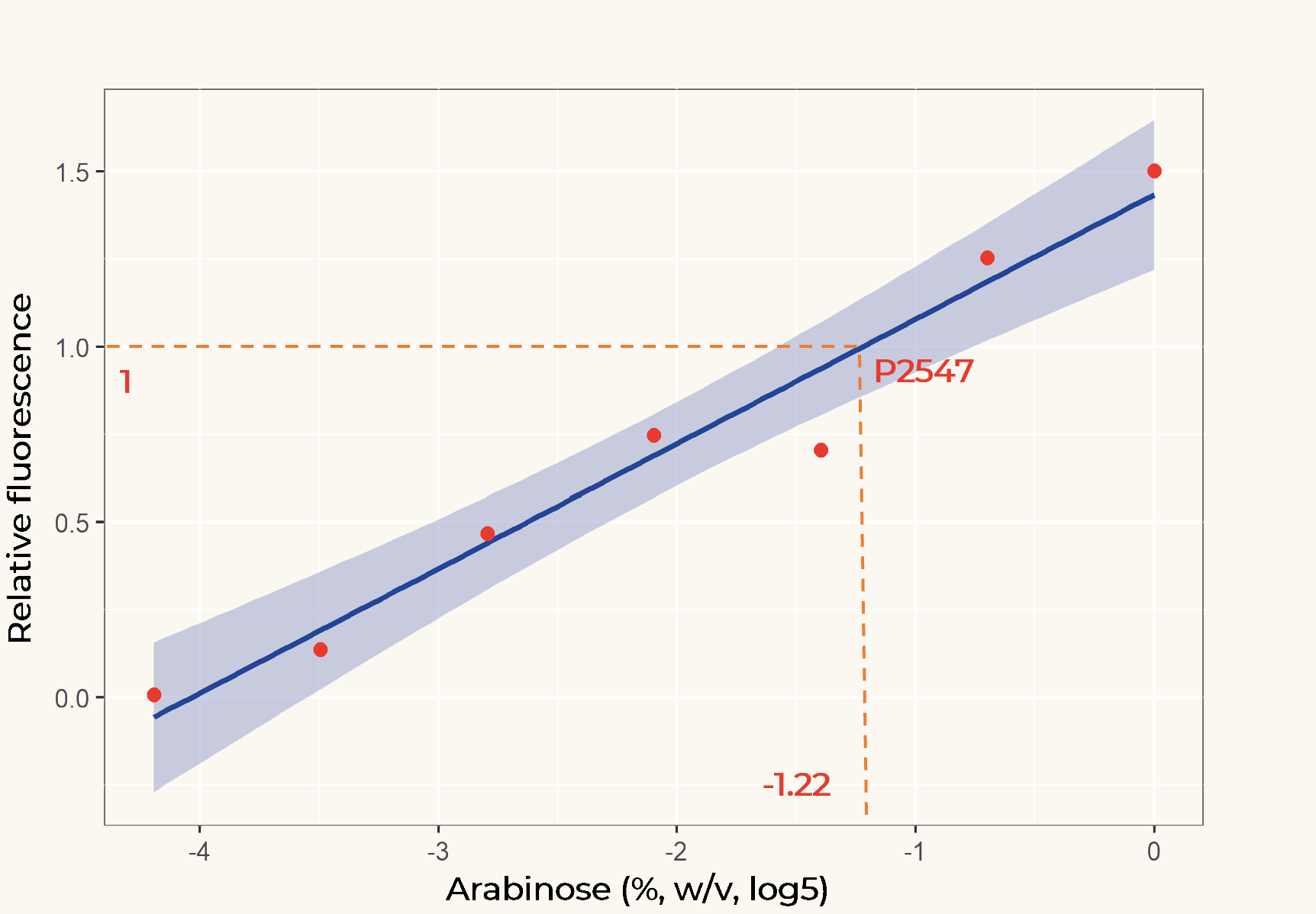
The results of the plate reader assay show that the inducible promoter pBAD gave varying levels of expression depending on L-arabinose concentration and constitutive promoter p2547 gave constant expression. At higher concentrations, the expression of pBAD is higher than the activity of p2547, and it is lower at concentrations approaching zero. We used this data to fit a linear regression model to determine at which concentration the level of pBAD equals p2547 expression (Fig. 2).
Figure 2. Linear regression of arabinose concentration relative fluorescence of pBAD compared with p2547. The orange dash lines showed how p2547 related to pBAD transcriptional activity at the arabinose concentration of 0.14% (log5-1.22).
The linear regression model showed that at a L-arabinose concentration of 0.14% (w/v), the expression of genes under pBAD is the same as under p2547.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]