Difference between revisions of "Part:BBa K3895003"
| (19 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
KerAVDZ50 is an extracellular serine thiol alkaline protease from the ''Actinomadura viridilutea'' strain, which can be isolated from the Algerian fishing port [1]. As a thermo-alkaline keratinases, it can function at a pH range of 7–12 and a temperature range of 35–80 °C. | KerAVDZ50 is an extracellular serine thiol alkaline protease from the ''Actinomadura viridilutea'' strain, which can be isolated from the Algerian fishing port [1]. As a thermo-alkaline keratinases, it can function at a pH range of 7–12 and a temperature range of 35–80 °C. | ||
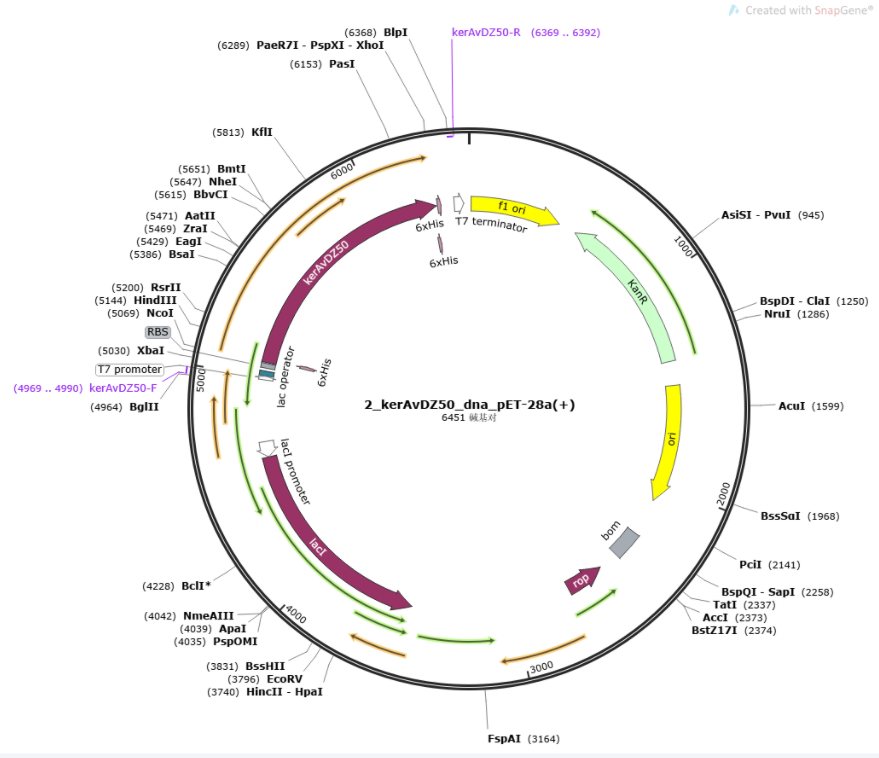
| − | 6x His-tags were added to both sides of the sequence for purification, and constructed into PET-28a(+) (<partinfo>K3895007</partinfo>). The length of KerAVDZ50 is | + | |
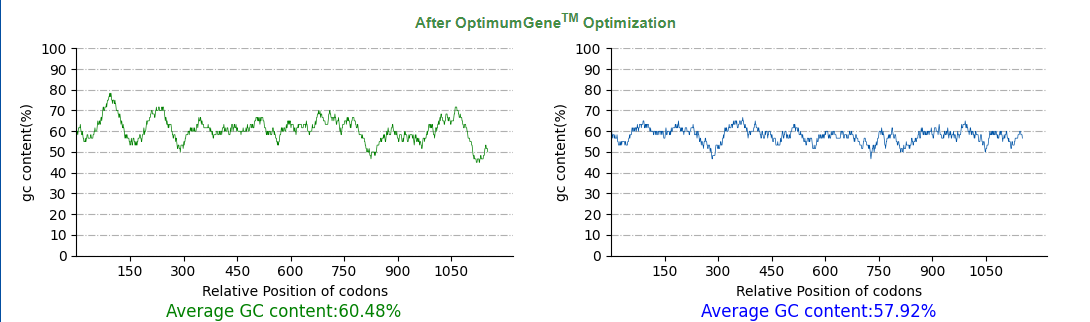
| + | [[File:T--SZ SHD--Z50O.png|900px|center]] | ||
| + | |||
| + | |||
| + | 6x His-tags were added to both sides of the sequence for purification, and constructed into PET-28a(+) (<partinfo>K3895007</partinfo>). The length of KerAVDZ50 is 1218 bp. | ||
| + | |||
{|border=0 width="90%" align="center" | {|border=0 width="90%" align="center" | ||
|-align="center" | |-align="center" | ||
| − | |[[File:T--SZ SHD--Z50plasmid.jpg| | + | |[[File:T--SZ SHD--Z50plasmid.jpg|300px|thumb|center|'''Figure 1.''' Plasmid construction of KerAVDZ50.]] |
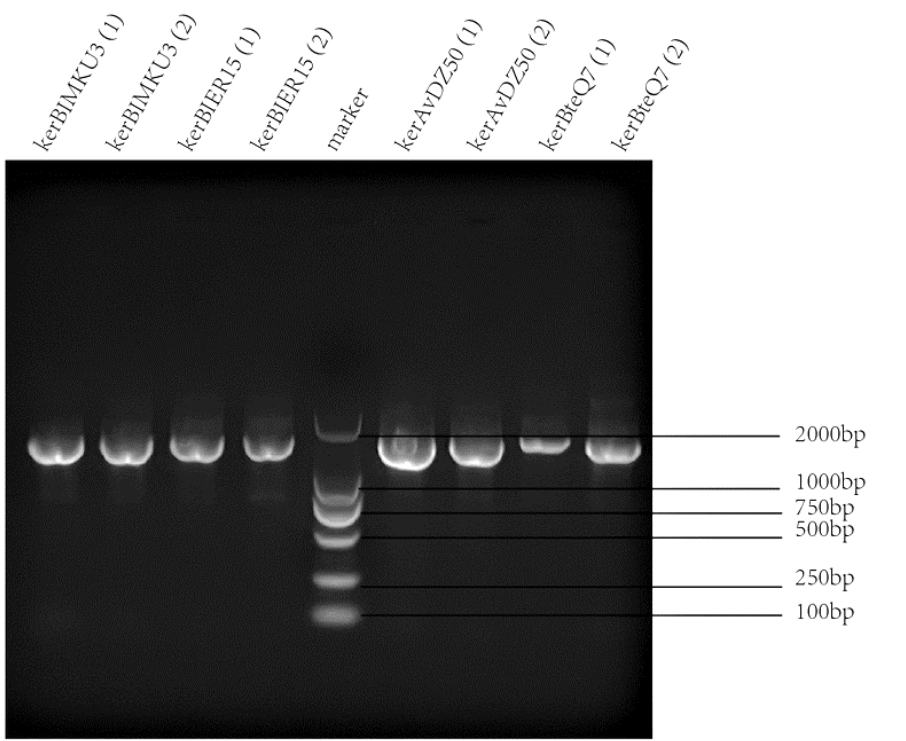
| − | |[[File:T--SZ SHD--PCR2.png| | + | |[[File:T--SZ SHD--PCR2.png|300px|thumb|center|''' Figure 2.''' PCR and gel electrophoresis of KerAVDZ50.]] |
|} | |} | ||
{|border=1 width="90%" align="center" | {|border=1 width="90%" align="center" | ||
|- | |- | ||
| − | !width="20%" style="background:#CCCCFF"|Name of | + | !width="20%" style="background:#CCCCFF"|Name of keratinase |
!width="20%"|IPTG concentration/mM | !width="20%"|IPTG concentration/mM | ||
!width="20%"|Temperature/°C | !width="20%"|Temperature/°C | ||
| Line 28: | Line 33: | ||
According to the amino acid sequences in the enzyme active sites and associated catalytic mechanisms, proteases can be classified into seven broad groups: serine, cysteine, threonine, aspartic and glutamic proteases, metalloproteases and asparagine peptide lyases. According to the nature of their active site, keratinases belong to serine- and metalloproteases or serine metalloproteases. Moreover, the enzymatic degradation of keratin is a multistage process that requires the following steps: adsorption of the keratinases to the surface of macromolecule by electrostatic and hydrophobic interactions, followed by catalytic action [2]. | According to the amino acid sequences in the enzyme active sites and associated catalytic mechanisms, proteases can be classified into seven broad groups: serine, cysteine, threonine, aspartic and glutamic proteases, metalloproteases and asparagine peptide lyases. According to the nature of their active site, keratinases belong to serine- and metalloproteases or serine metalloproteases. Moreover, the enzymatic degradation of keratin is a multistage process that requires the following steps: adsorption of the keratinases to the surface of macromolecule by electrostatic and hydrophobic interactions, followed by catalytic action [2]. | ||
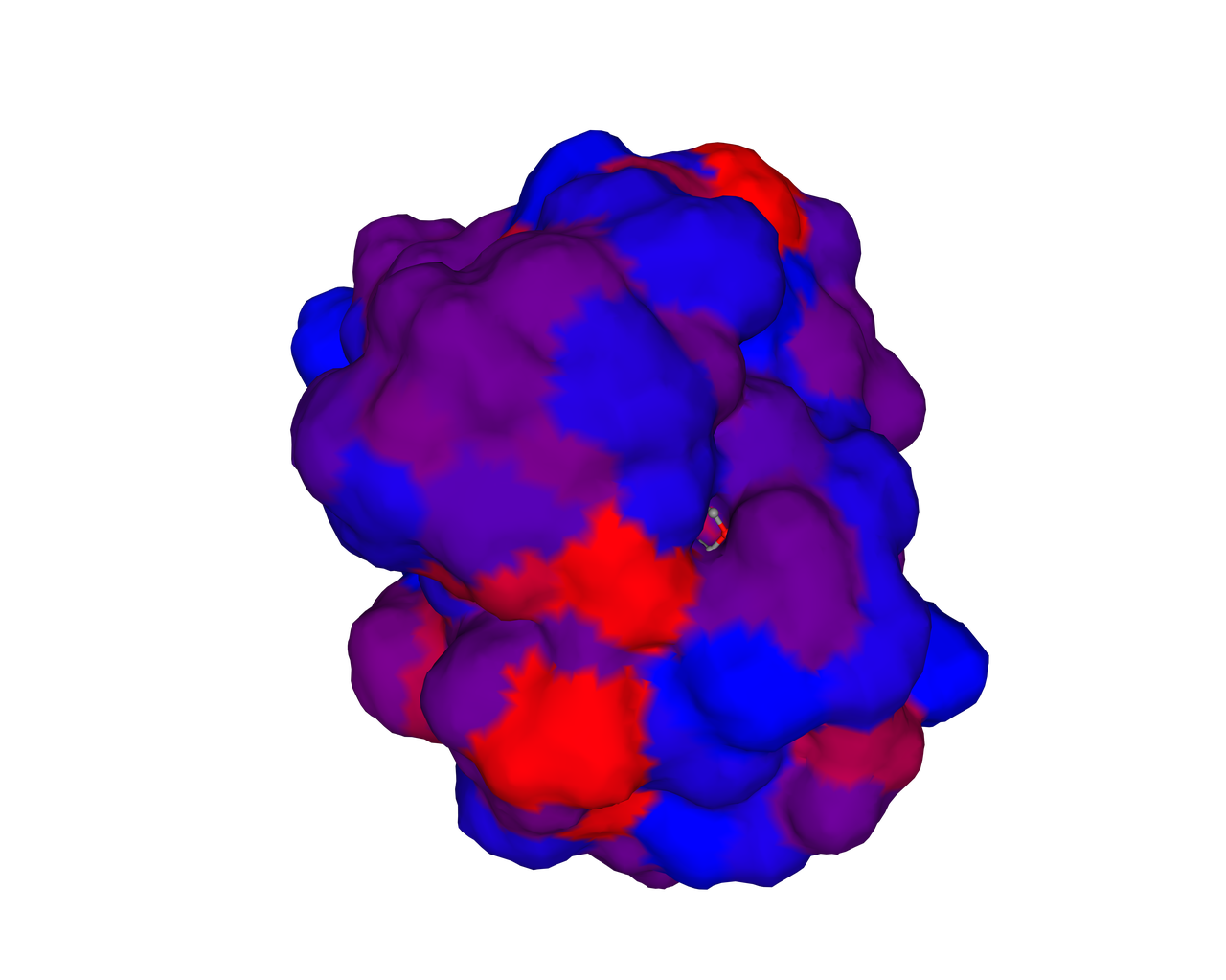
Hence we used Swiss Model to predict their hydrophobic region and serine groups. | Hence we used Swiss Model to predict their hydrophobic region and serine groups. | ||
| + | |||
| + | |||
{|border=0 width="90%" align="center" | {|border=0 width="90%" align="center" | ||
|-align="center" | |-align="center" | ||
| − | |[[File:T--SZ SHD--Z50.1.gif| | + | |[[File:T--SZ SHD--Z50.1.gif|300px|thumb|center|'''Figure 1.''' Swiss Model showing serine residues of KerAVDZ50.]] |
| − | |[[File:T--SZ SHD--Z50.2.png| | + | |[[File:T--SZ SHD--Z50.2.png|300px|thumb|center|''' Figure 2.''' Swiss Model showing hydrophobisity of KerAVDZ50.]] |
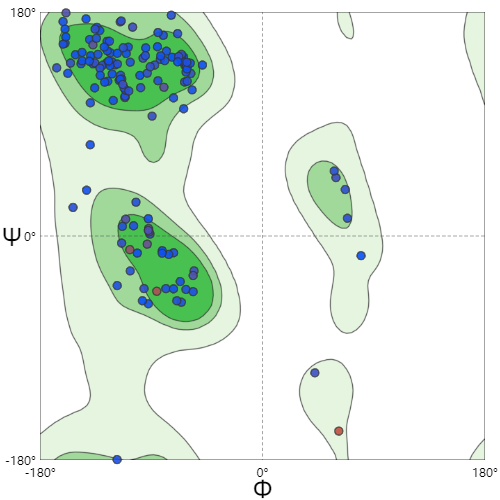
| − | |[[File:T--SZ SHD--Z50R.jpg| | + | |[[File:T--SZ SHD--Z50R.jpg|300px|thumb|center|''' Figure 3.''' Ramachandran Plot of KerAVDZ50.Z50 Ramachandran Favoured is 95.14%]] |
|} | |} | ||
| Line 53: | Line 60: | ||
===References=== | ===References=== | ||
Biochemical and molecular characterization of new keratinoytic protease from Actinomadura viridilutea DZ50. (n.d.). International Journal of Biological Macromolecules, 92, 299–315. https://doi.org/10.1016/j.ijbiomac.2016.07.009 | Biochemical and molecular characterization of new keratinoytic protease from Actinomadura viridilutea DZ50. (n.d.). International Journal of Biological Macromolecules, 92, 299–315. https://doi.org/10.1016/j.ijbiomac.2016.07.009 | ||
| + | |||
Vidmar, Beti, and Maša Vodovnik. “Microbial Keratinases: Enzymes with Promising Biotechnological Applications.” Food technology and biotechnology vol. 56,3 (2018): 312-328. doi:10.17113/ftb.56.03.18.5658 | Vidmar, Beti, and Maša Vodovnik. “Microbial Keratinases: Enzymes with Promising Biotechnological Applications.” Food technology and biotechnology vol. 56,3 (2018): 312-328. doi:10.17113/ftb.56.03.18.5658 | ||
Latest revision as of 08:01, 11 October 2021
Keratinase kerAvDZ50
KerAVDZ50 is an extracellular serine thiol alkaline protease from the Actinomadura viridilutea strain, which can be isolated from the Algerian fishing port [1]. As a thermo-alkaline keratinases, it can function at a pH range of 7–12 and a temperature range of 35–80 °C.
6x His-tags were added to both sides of the sequence for purification, and constructed into PET-28a(+) (BBa_K3895007). The length of KerAVDZ50 is 1218 bp.
| Name of keratinase | IPTG concentration/mM | Temperature/°C | Time/h |
|---|---|---|---|
| KerAVDZ50 | 0.67139 | 16 | 12 |
Modeling
The biological activity of a protein is not only determined by the primary structure of the protein molecule but also closely related to its specific spatial structure. Elucidating the process of protein folding in functional and structural details will be of great significance to evaluate their keratinotic functions. According to the amino acid sequences in the enzyme active sites and associated catalytic mechanisms, proteases can be classified into seven broad groups: serine, cysteine, threonine, aspartic and glutamic proteases, metalloproteases and asparagine peptide lyases. According to the nature of their active site, keratinases belong to serine- and metalloproteases or serine metalloproteases. Moreover, the enzymatic degradation of keratin is a multistage process that requires the following steps: adsorption of the keratinases to the surface of macromolecule by electrostatic and hydrophobic interactions, followed by catalytic action [2]. Hence we used Swiss Model to predict their hydrophobic region and serine groups.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 577
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 382
Illegal AgeI site found at 1120 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 322
References
Biochemical and molecular characterization of new keratinoytic protease from Actinomadura viridilutea DZ50. (n.d.). International Journal of Biological Macromolecules, 92, 299–315. https://doi.org/10.1016/j.ijbiomac.2016.07.009
Vidmar, Beti, and Maša Vodovnik. “Microbial Keratinases: Enzymes with Promising Biotechnological Applications.” Food technology and biotechnology vol. 56,3 (2018): 312-328. doi:10.17113/ftb.56.03.18.5658