Difference between revisions of "Part:BBa K3887004"
NgSweeHong (Talk | contribs) (→Contribution: Tsinghua 2023 uses pCas to knock out ArgR gene) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3887004 short</partinfo> | <partinfo>BBa_K3887004 short</partinfo> | ||
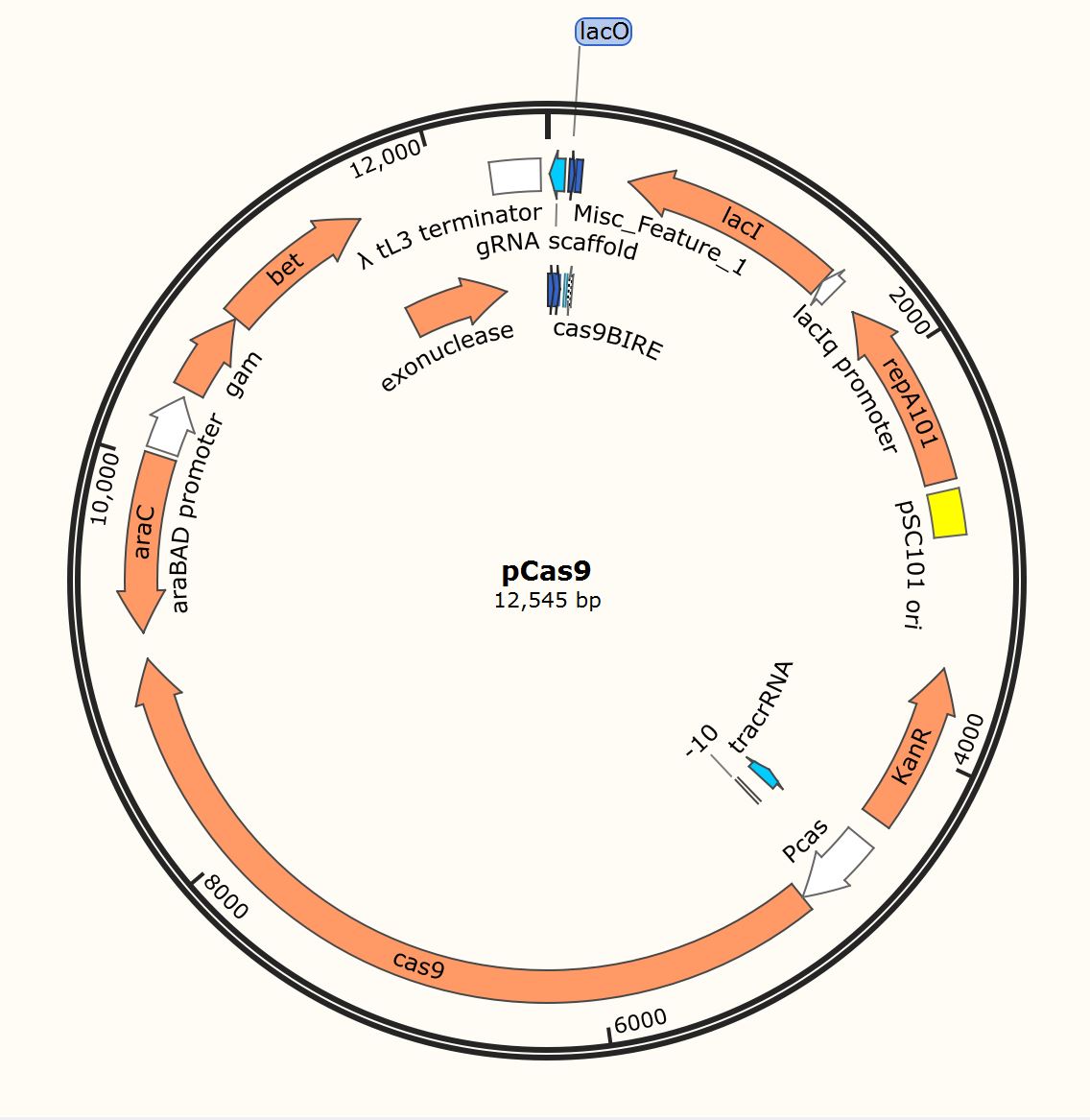
| − | Cas9 editing protein | + | pCas9 is a plamid containing Cas9 editing enzyme, used for gene modification at a specific location. Cas9 protein follows guide RNA to cut the two strands of DNA at a designated location, where strands of DNA can be edit (insertion, deletion,mutation). |
| + | In our project,we used this plasmid to knockout TnaA on <i>E. coli </i> BL21(DE3) genome. Below is the map of it. | ||
| + | [[File:T--BJU_China--Fig.1 pCas9-1.png|300px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure.1 Snapgene map of pCas9</center></b></p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 20: | ||
<partinfo>BBa_K3887004 parameters</partinfo> | <partinfo>BBa_K3887004 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Contribution: Tsinghua 2023 uses pCas to knock out ArgR gene=== | ||
| + | <p>PCas is one of the commonly used plasmids in gene knockout of Escherichia coli. Mainly including Cas9 expression elements, Red homologous recombination system, sgRNA-PMB1 (for pTargetF removal) and Rep101 (for pCas removal)</p> | ||
| + | |||
| + | <p>This year, Tsinghua 2023 successfully knocked out the ArgR gene in MG1655 using pTargetF/pCas. We adopted http://www.rgenome.net/cas-designer/notice Conduct N20 design. At the same time, we pioneered the simultaneous transformation of homologous arms, pCAS, and pTargetF into Escherichia coli, and successfully achieved gene knockout (see figure below). This innovation greatly reduces the operational complexity and experimental cycle of the prokaryotic gene knockout system.</p> | ||
| + | |||
| + | <html> | ||
| + | <div class="center"> | ||
| + | <div class="thumb tnone"> | ||
| + | <div class="thumbinner" style="width:50%;"> | ||
| + | <a href="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-1.png" class="image"> | ||
| + | <img alt="" src="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-1.png" width="100%" height=auto class="thumbimage" /></a> <div class="thumbcaption"> | ||
| + | <div class="magnify"> | ||
| + | <a href="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-1.png" class="internal" title="Enlarge"></a> | ||
| + | </div> | ||
| + | <b></b> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <div class="center"> | ||
| + | <div class="thumb tnone"> | ||
| + | <div class="thumbinner" style="width:50%;"> | ||
| + | <a href="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-2.png" class="image"> | ||
| + | <img alt="" src="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-2.png" width="100%" height=auto class="thumbimage" /></a> <div class="thumbcaption"> | ||
| + | <div class="magnify"> | ||
| + | <a href="https://static.igem.wiki/teams/4634/wiki/parts-registry/ptargetfpcas-2.png" class="internal" title="Enlarge"></a> | ||
| + | </div> | ||
| + | <b></b> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 18:18, 11 October 2023
pCas
pCas9 is a plamid containing Cas9 editing enzyme, used for gene modification at a specific location. Cas9 protein follows guide RNA to cut the two strands of DNA at a designated location, where strands of DNA can be edit (insertion, deletion,mutation). In our project,we used this plasmid to knockout TnaA on E. coli BL21(DE3) genome. Below is the map of it.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 6015
Illegal NotI site found at 3034 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 8294
Illegal BamHI site found at 10296
Illegal XhoI site found at 4353 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 10131
Illegal AgeI site found at 11788 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 10113
Contribution: Tsinghua 2023 uses pCas to knock out ArgR gene
PCas is one of the commonly used plasmids in gene knockout of Escherichia coli. Mainly including Cas9 expression elements, Red homologous recombination system, sgRNA-PMB1 (for pTargetF removal) and Rep101 (for pCas removal)
This year, Tsinghua 2023 successfully knocked out the ArgR gene in MG1655 using pTargetF/pCas. We adopted http://www.rgenome.net/cas-designer/notice Conduct N20 design. At the same time, we pioneered the simultaneous transformation of homologous arms, pCAS, and pTargetF into Escherichia coli, and successfully achieved gene knockout (see figure below). This innovation greatly reduces the operational complexity and experimental cycle of the prokaryotic gene knockout system.