Difference between revisions of "Part:BBa K2965021"
(→Improvement) |
|||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 31: | Line 31: | ||
[[File:T--ShanghaiTech_China--ascpfparts1WB22.png|center|500px|thumb|'''Figure 6. AsCpf1 western blot with histag.''']] | [[File:T--ShanghaiTech_China--ascpfparts1WB22.png|center|500px|thumb|'''Figure 6. AsCpf1 western blot with histag.''']] | ||
We got our wide-type AsCas12a protein (link:<bbpart>BBa_K3454037</bbpart>) from <bbpart>BBa_K2965021</bbpart>. For higher expression yield in E. coli, we did codon optimization on the original coding sequence. Although the DNA sequences was changed, the amino acid sequence remained the same, so the protein was not changed after codon optimization. Then we introduced the selected mutations into WT plasmid through circular PCR reaction in the order of S542R, K548V, N552R and E174R, which is <bbpart>BBa_K3454038</bbpart>, <bbpart>BBa_K3454039</bbpart> and <bbpart>BBa_K3454040</bbpart>, respectively. And for composite part with Histag and MBP tag: <bbpart>BBa_K3454041</bbpart>, <bbpart>BBa_K3454042</bbpart> and <bbpart>BBa_K3454043</bbpart>. As a result, we successfully extracted three mutant proteins —— AsCas12a-R (<bbpart>BBa_K3454038</bbpart>), AsCas12a-RVR (<bbpart>BBa_K3454039</bbpart>) and AsCas12a-RVRR (<bbpart>BBa_K3454040</bbpart>). | We got our wide-type AsCas12a protein (link:<bbpart>BBa_K3454037</bbpart>) from <bbpart>BBa_K2965021</bbpart>. For higher expression yield in E. coli, we did codon optimization on the original coding sequence. Although the DNA sequences was changed, the amino acid sequence remained the same, so the protein was not changed after codon optimization. Then we introduced the selected mutations into WT plasmid through circular PCR reaction in the order of S542R, K548V, N552R and E174R, which is <bbpart>BBa_K3454038</bbpart>, <bbpart>BBa_K3454039</bbpart> and <bbpart>BBa_K3454040</bbpart>, respectively. And for composite part with Histag and MBP tag: <bbpart>BBa_K3454041</bbpart>, <bbpart>BBa_K3454042</bbpart> and <bbpart>BBa_K3454043</bbpart>. As a result, we successfully extracted three mutant proteins —— AsCas12a-R (<bbpart>BBa_K3454038</bbpart>), AsCas12a-RVR (<bbpart>BBa_K3454039</bbpart>) and AsCas12a-RVRR (<bbpart>BBa_K3454040</bbpart>). | ||
| − | For more detail, see [https://2020.igem.org/Team:ShanghaiTech_China/Improvement] | + | For more detail, see our improvement wiki page:[https://2020.igem.org/Team:ShanghaiTech_China/Improvement]. |
====Reference==== | ====Reference==== | ||
| Line 48: | Line 48: | ||
<partinfo>BBa_K2965021 parameters</partinfo> | <partinfo>BBa_K2965021 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Effect of ions on AsCas12a’s collateral cleavage activity=== | ||
| + | ====iGEM Thrace 2021==== | ||
| + | AsCas12a’s activity is modified in the presence of certain ions. In particular, AsCas12a is able to degrade DNA substrate in the presence of Mg2+ and/orMn2+. Mg2+ is required for a crRNA free cleavage activity of DNA substrate. Furthermore, in the presence of Mg2+ buffer, AsCas12a displays a preferred endonuclease activity for ssDNA rather than dsDNA. | ||
| + | For circular ssDNA, AsCas12a enzyme has the same activity for the same period of time, no matter which kind of metal ion (Mg2+ or Mn2+) is used. Another ion that promotes the cleavage of ssDNA from AsCas12a is Co2+. | ||
| + | |||
| + | ====References==== | ||
| + | Li, B., Yan, J., Zhang, Y., Li, W., Zeng, C., Zhao, W., Hou, X., Zhang, C., & Dong, Y. (2020). CRISPR-CAS12A possesses unconventional DNase activity that can be inactivated by synthetic oligonucleotides. Molecular Therapy - Nucleic Acids, 19, 1043–1052. https://doi.org/10.1016/j.omtn.2019.12.038 | ||
| + | |||
| + | ===Trans-cleavage activity of AsCas12a=== | ||
| + | ====iGEM NTU-Singapore 2021==== | ||
| + | Based on a paper published by a research team in Nanyang Technological University (NTU), a test was conducted to examine the sensitivity of various Cas12 enzymes[1]. AsCas12a is able to detect for COVID19 RNA at the S2 site when the guide RNA perfectly matches the sequence. In the experiment, the gRNA sequence that was used is ACTCCTGGTGATTCTTCTT. Based on figure a, AsCas12a is able to specifically detect for COVID19 as there is no cross-reactivity for SARS-CoV or MERS-CoV. At 37°C, the fluorescence signal for SARS-CoV-2 is twice as high as compared to the fluorescence signal at 24°C. | ||
| + | |||
| + | In the paper, 10 different guide RNAs were made to test for mismatched activity of the different Cas enzyme. It was identified that AsCas12a produces minimal fluorescence reading when there is a mismatch of 1 nucleotide. This shows that AsCas12a is sensitive to a single nucleotide discrimination. This is also true for the LBCas12a. | ||
| + | |||
| + | [[File:T--NTU-Singapore--Contribution1.png|center|700px|thumb|'''a.Intensity of the fluorescence produced for different Cas12a enzymes using single S2 gRNA sequence. b. Heatmap showing the intensity of the fluorescence produced after trans-cleavage assay was performed at 37 °C. 1 - highest fluorescence intensity measured & 0 - lowest fluorescence intensity''']] | ||
| + | |||
| + | ====References==== | ||
| + | Ooi, K. H., Liu, M. M., Tay, J. W., Teo, S. Y., Kaewsapsak, P., Jin, S., Lee, C. K., Hou, J., Maurer-Stroh, S., Lin, W., Yan, B., Yan, G., Gao, Y.-G., & Tan, M. H. (2021). An engineered CRISPR-CAS12A variant and DNA-RNA hybrid guides enable robust and rapid covid-19 testing. Nature Communications, 12(1). https://doi.org/10.1038/s41467-021-21996-6 | ||
Latest revision as of 19:47, 21 October 2021
AsCas12a (AsCpf1)
AsCas12a (AsCpf1) comes from Acidaminococcus sp. BV3L6, it is a programmable DNA endonuclease guided by a single CRISPR RNA (crRNA). Targeting requires a crRNA complementary to the target site as well as a 5' TTTN protospacer adjacent motif (PAM) on the DNA strand opposite the target sequence. Once it recognizes its target, it will be activated and exhibits a “collateral effects” of promiscuous DNase activity.
Usage and Biology
AsCas12a RNPs bind to DNA and start to move along it by a hopping mechanism. When the AsCas12a RNPs encounter a potential target site containing a PAM sequence, they begin to initiate R-loop formation by forming base-pair hybrids between the crRNA and the target DNA strand. If sufficient matched nucleotides exist in the PAM-proximal region, the R-loop formation will be stable. After the stable R-loop conformation is formed, AsCas12a, via the activity of its RuvC domain, first cleaves the non-target DNA strand and then cleaves the opposite, target DNA strand, regardless of the presence of a PAM sequence. Finally, AsCas12a rapidly releases the PAM-distal DNA fragment but continues to hold the PAM-proximal portion of DNA[1].
Characterization
Results
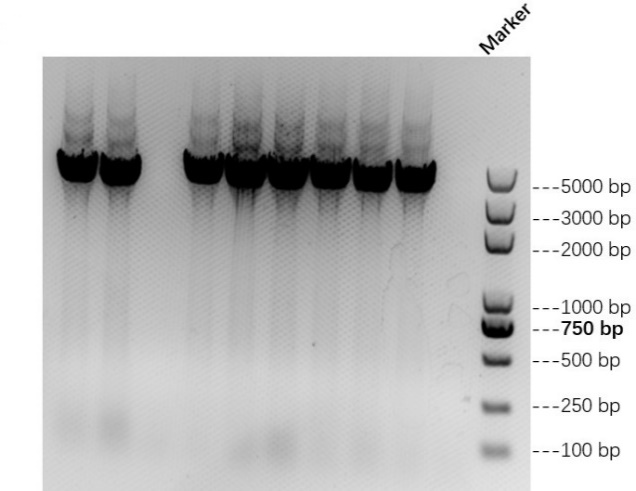
We link AsCas12a fragment and pET-28a(+) plasmid to form AbCas12a expression plasmid with His tag. Then we transfer the recombinant plasmid into E. coli BL21 and test it by colony PCR. Result is shown in Figure 1 below. We also verify it by TSINGKE Biological Technology Institute sequencing.
We induce AsCas12a expression and purify it by gravity-flow column with Ni-NTA Sefinose(TM) Resin. SDS-PAGE and Western Blot is used to characterize protein purification as shown in Figure 2 and Figure 3.
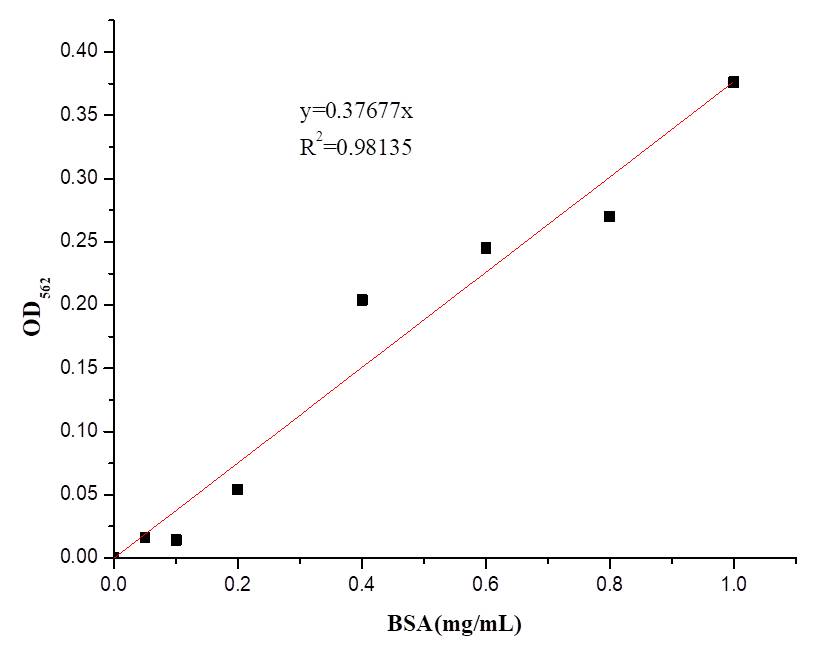
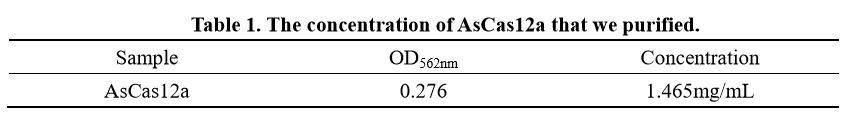
We also determine the protein concentration with BCA kit.
Finally, we test the activity of AsCas12a by detecting the fluorescence. A kind of 5bp single strand DNA with 5'-FAM and 3'-TAMRA is used as reporters in which the fluorescence of FAM can be quenched by TAMRA to display its DNA endonuclease activity. When AsCas12a is activated and cuts reporters, the cleavage of reporters will enable FAM to emit green fluorescence and neutralize the red fluorescence of TAMRA to be yellow. The extracted AsCas12a has ability to cut DNA compared to the control group and significantly higher activity compared to the bought AsCas12a.
Improvement
iGEM Shanghaitech_China 2020 has improvevd AsCas12a (link:BBa_K3454046) in the way that two tags are added, namely MBP tag and His tag. MBP tag is maltose binding protein, which helps Cas12a expression, dissolution and folding in correct conformation. His tag can help purification Cas12a through Ni-NTA beads.
We got our wide-type AsCas12a protein (link:BBa_K3454037) from BBa_K2965021. For higher expression yield in E. coli, we did codon optimization on the original coding sequence. Although the DNA sequences was changed, the amino acid sequence remained the same, so the protein was not changed after codon optimization. Then we introduced the selected mutations into WT plasmid through circular PCR reaction in the order of S542R, K548V, N552R and E174R, which is BBa_K3454038, BBa_K3454039 and BBa_K3454040, respectively. And for composite part with Histag and MBP tag: BBa_K3454041, BBa_K3454042 and BBa_K3454043. As a result, we successfully extracted three mutant proteins —— AsCas12a-R (BBa_K3454038), AsCas12a-RVR (BBa_K3454039) and AsCas12a-RVRR (BBa_K3454040). For more detail, see our improvement wiki page:[1].
Reference
[1] Jeon Y, Choi YH, Jang Y, Yu J, Goo J, Lee G, et al. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nature Communications. 2018;9(1):2777-11.
[2] Mohanraju, P., Oost, J. v., Jinek, M. and Swarts, D. C. (2018). Heterologous Expression and Purification of the CRISPR-Cas12a/Cpf1 Protein. Bio-protocol 8(9): e2842. DOI: 10.21769/BioProtoc.2842.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 380
Illegal BglII site found at 1922
Illegal BglII site found at 2231 - 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865
Illegal NgoMIV site found at 3466 - 1000COMPATIBLE WITH RFC[1000]
Effect of ions on AsCas12a’s collateral cleavage activity
iGEM Thrace 2021
AsCas12a’s activity is modified in the presence of certain ions. In particular, AsCas12a is able to degrade DNA substrate in the presence of Mg2+ and/orMn2+. Mg2+ is required for a crRNA free cleavage activity of DNA substrate. Furthermore, in the presence of Mg2+ buffer, AsCas12a displays a preferred endonuclease activity for ssDNA rather than dsDNA. For circular ssDNA, AsCas12a enzyme has the same activity for the same period of time, no matter which kind of metal ion (Mg2+ or Mn2+) is used. Another ion that promotes the cleavage of ssDNA from AsCas12a is Co2+.
References
Li, B., Yan, J., Zhang, Y., Li, W., Zeng, C., Zhao, W., Hou, X., Zhang, C., & Dong, Y. (2020). CRISPR-CAS12A possesses unconventional DNase activity that can be inactivated by synthetic oligonucleotides. Molecular Therapy - Nucleic Acids, 19, 1043–1052. https://doi.org/10.1016/j.omtn.2019.12.038
Trans-cleavage activity of AsCas12a
iGEM NTU-Singapore 2021
Based on a paper published by a research team in Nanyang Technological University (NTU), a test was conducted to examine the sensitivity of various Cas12 enzymes[1]. AsCas12a is able to detect for COVID19 RNA at the S2 site when the guide RNA perfectly matches the sequence. In the experiment, the gRNA sequence that was used is ACTCCTGGTGATTCTTCTT. Based on figure a, AsCas12a is able to specifically detect for COVID19 as there is no cross-reactivity for SARS-CoV or MERS-CoV. At 37°C, the fluorescence signal for SARS-CoV-2 is twice as high as compared to the fluorescence signal at 24°C.
In the paper, 10 different guide RNAs were made to test for mismatched activity of the different Cas enzyme. It was identified that AsCas12a produces minimal fluorescence reading when there is a mismatch of 1 nucleotide. This shows that AsCas12a is sensitive to a single nucleotide discrimination. This is also true for the LBCas12a.
References
Ooi, K. H., Liu, M. M., Tay, J. W., Teo, S. Y., Kaewsapsak, P., Jin, S., Lee, C. K., Hou, J., Maurer-Stroh, S., Lin, W., Yan, B., Yan, G., Gao, Y.-G., & Tan, M. H. (2021). An engineered CRISPR-CAS12A variant and DNA-RNA hybrid guides enable robust and rapid covid-19 testing. Nature Communications, 12(1). https://doi.org/10.1038/s41467-021-21996-6