Difference between revisions of "Part:BBa K3588014"
(→design) |
|||
| (23 intermediate revisions by the same user not shown) | |||
| Line 11: | Line 11: | ||
Our design avoids this issue and means that the kill switch can be employed in any and all situations. | Our design avoids this issue and means that the kill switch can be employed in any and all situations. | ||
| − | === | + | ===Design=== |
When approaching our design understood that we had to be able to compare in a quantifiable way the amount we would be expressing each protein. There are two parts of a circuit that effect expression: the promoter and the RBS. | When approaching our design understood that we had to be able to compare in a quantifiable way the amount we would be expressing each protein. There are two parts of a circuit that effect expression: the promoter and the RBS. | ||
| Line 39: | Line 39: | ||
| + | |||
| + | |||
| + | |||
| + | [[File:T--OFFICIAL CLS CLSG UK--kill switch promoter strengths.jpeg|600px|left|thumb|Calculations for promoter strengths]] | ||
| Line 47: | Line 51: | ||
compiling these tables gives: | compiling these tables gives: | ||
| − | |||
| Line 55: | Line 58: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 69: | Line 65: | ||
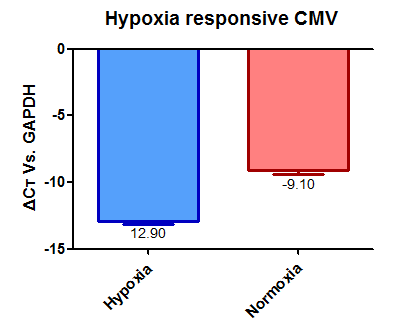
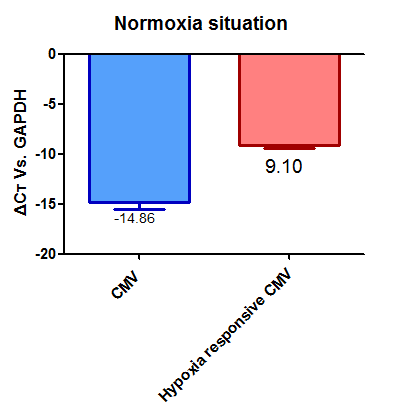
| + | From these we can say that promoter [[part:BBa_K1720002]] has a relative strengths of 0.60 in normoxia and 0.87 in hypoxia. | ||
| + | While promoter [[part:BBa_K747096]] has relative strengths of 1.00 in normoxia and hypoxia. | ||
| Line 93: | Line 91: | ||
| + | Ribosomes: | ||
| + | Of course, ribosome binding strength is also a significant part of protein expression, so a paper on the relative strengths of RBS was also used. | ||
| + | [[File:T--OFFICIAL CLS CLSG RBS strrengths.png|500px|left|thumb|Relative strengths of RBS on the registry<ref>https://digitalcommons.usu.edu/cgi/viewcontent.cgi?article=1873&context=etd</ref>]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | From this paper it says that the relative strength of | ||
| + | [[part:B0034]] = 1.748 | ||
| + | [[part:B0031]] = 1.101 | ||
| + | Therefore, it appears that [[part:B0031]] is 63% as efficient as [[part:B0034]] | ||
| Line 113: | Line 123: | ||
| + | Combining it all together gives us: | ||
| + | [[File:T--OFFICIAL CLS CLSG UK--kill switch in normoxia.png|center|900px|thumb|Full kill switch in normoxic conditions]] | ||
| + | [[File:T--OFFICIAL CLS CLSG UK--kill switch in hypoxia.png|center|900px|thumb|Full kill switch in hypoxic conditions]] | ||
| + | We can then see that while in normoxic conditions the kill switch would have no significant effect upon the mortality, metabolism or efficiency of the cell. Meanwhile as soon as hypoxic conditions are met the amount of MazF produced will increase and so the cell will undergo apoptosis rapidly. | ||
| Line 124: | Line 138: | ||
<partinfo>BBa_K3588014 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3588014 SequenceAndFeatures</partinfo> | ||
| + | ===Synthesis=== | ||
| + | When trying to order this insert from IDT the insert as a whole was too complex for synthesis. Given this we ran codon optimisation and we intended to order the two circuits separately and then run a digestion and ligation to create one insert. So, the code above is the codon optimised version of the original parts. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 03:05, 28 October 2020
MazE-MazF Hypoxia induced kill switch
MazF is under control of the promoter BBa_K1720002, upregulating in anoxic conditions. MazE is under control of the constitutive promoter BBa_K747096, downregulating in anoxic conditions. In anoxic conditions, there should be excess MazF, leading to apoptotic cell death.
Our Improvements
This kill switch is a significant improvement on other kill switches that employ the same toxin-antitoxin system. By controlling the expression of each of the proteins with different promoters we have avoided the issues that occurred with previous when emplying this toxin-antitoxin in a kill switch. Previously the activation and effectiveness of the kill switch is reliant on the differing cellular breakdown rates of the compounds. As has been established through lab work previously, this means that no cells, no matter the conditions they are in, can survive for significant periods of time. While this doesn’t pose an issue for containment it does pose an issue for the sustainability of the kill switch as well as the sustainability of any long term bioengineering based solution that hopes to use their system. Our design avoids this issue and means that the kill switch can be employed in any and all situations.
Design
When approaching our design understood that we had to be able to compare in a quantifiable way the amount we would be expressing each protein. There are two parts of a circuit that effect expression: the promoter and the RBS.
Promoters: We knew that we wanted to have our kill switch triggered by hypoxia so we used the part:BBa_K1720002 promoter and the CMV promoter part:BBa_K747096. These two promoters have been characterised against each other.
compiling these tables gives:
From these we can say that promoter part:BBa_K1720002 has a relative strengths of 0.60 in normoxia and 0.87 in hypoxia.
While promoter part:BBa_K747096 has relative strengths of 1.00 in normoxia and hypoxia.
Ribosomes:
Of course, ribosome binding strength is also a significant part of protein expression, so a paper on the relative strengths of RBS was also used.

From this paper it says that the relative strength of
part:B0034 = 1.748
part:B0031 = 1.101
Therefore, it appears that part:B0031 is 63% as efficient as part:B0034
Combining it all together gives us:
We can then see that while in normoxic conditions the kill switch would have no significant effect upon the mortality, metabolism or efficiency of the cell. Meanwhile as soon as hypoxic conditions are met the amount of MazF produced will increase and so the cell will undergo apoptosis rapidly.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 1043
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 1043
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 576
Illegal BamHI site found at 655 - 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 1043
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 1043
- 1000COMPATIBLE WITH RFC[1000]
Synthesis
When trying to order this insert from IDT the insert as a whole was too complex for synthesis. Given this we ran codon optimisation and we intended to order the two circuits separately and then run a digestion and ligation to create one insert. So, the code above is the codon optimised version of the original parts.