Difference between revisions of "Part:BBa K3645012"
Teddy Huang (Talk | contribs) |
Teddy Huang (Talk | contribs) (→Peking iGEM 2020's doncumentation) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == Peking iGEM 2020's documentation == | |
| − | + | <p>David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand | |
| − | + | their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically | |
| − | + | impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes | |
| − | + | from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by | |
| − | + | adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies | |
| − | + | the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the | |
| − | + | deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may | |
| − | + | be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna | |
| − | + | Bachle) </p> | |
| − | + | <p>We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After | |
| − | + | multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as | |
| − | + | follows:</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | == Peking iGEM 2020's | + | |
| − | <p>David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna Bachle) </p><p>We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as follows:</p> | + | |
<h1>ABE</h1> | <h1>ABE</h1> | ||
| − | + | https://2020.igem.org/wiki/images/f/f2/T--Peking--ABE-4.jpeg | |
| − | + | <h6>(Gaudelli et al., 2020.)</h6><br> | |
| − | <p>Gaudelli et al. have successfully developed an adenosine deaminase, which can act on DNA for adenine base editing. They first created a defective chloramphenicol resistance gene (CamR) by introducing a point mutation (H193Y). Reversal of this mutation by adenine base editor will restore antibiotic resistance. To find such a protein, they created a mutant library of E.coli tRNA adenosine deaminase (ecTadA), fused it with dcas9, and transformed it into E.coli containing the defective CamR gene. Screening of viable colonies and subsequent rounds of evolution and engineering produced a mutant TadA (TadA *), which accepted DNA as a substrate satisfactorily. </p><p> | + | <p>Gaudelli et al. have successfully developed an adenosine deaminase, which can act on DNA for adenine base editing. |
| − | The artificially evolved adenosine deaminase catalyzes the transformation of target "A" into "I" (inosine), which is regarded as "G" by cell polymerase. Subsequently, a primitive genome A•T base pair was transformed into a G•C base pair. Since inosine excision repair is not as active as uracil excision, ABE does not require any additional inhibitor proteins, such as UGI in CBE. | + | They first created a defective chloramphenicol resistance gene (CamR) by introducing a point mutation (H193Y). |
| + | Reversal of this mutation by adenine base editor will restore antibiotic resistance. To find such a protein, they | ||
| + | created a mutant library of E.coli tRNA adenosine deaminase (ecTadA), fused it with dcas9, and transformed it into | ||
| + | E.coli containing the defective CamR gene. Screening of viable colonies and subsequent rounds of evolution and | ||
| + | engineering produced a mutant TadA (TadA *), which accepted DNA as a substrate satisfactorily. </p> | ||
| + | <p> | ||
| + | The artificially evolved adenosine deaminase catalyzes the transformation of target "A" into "I" (inosine), which is | ||
| + | regarded as "G" by cell polymerase. Subsequently, a primitive genome A•T base pair was transformed into a G•C base | ||
| + | pair. Since inosine excision repair is not as active as uracil excision, ABE does not require any additional | ||
| + | inhibitor proteins, such as UGI in CBE. | ||
</p> | </p> | ||
| − | + | https://2020.igem.org/wiki/images/d/d8/T--Peking--ABE-1.png | |
| − | + | https://2020.igem.org/wiki/images/f/f7/T--Peking--ABE-2.png | |
| − | + | https://2020.igem.org/wiki/images/f/f7/T--Peking--ABE-3.png | |
Latest revision as of 21:32, 27 October 2020
Peking iGEM 2020's documentation
David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna Bachle)
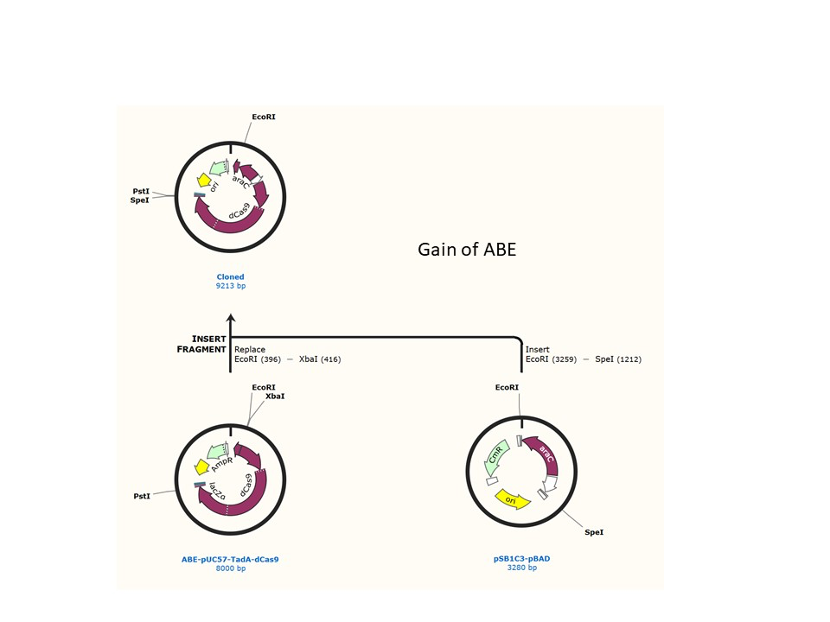
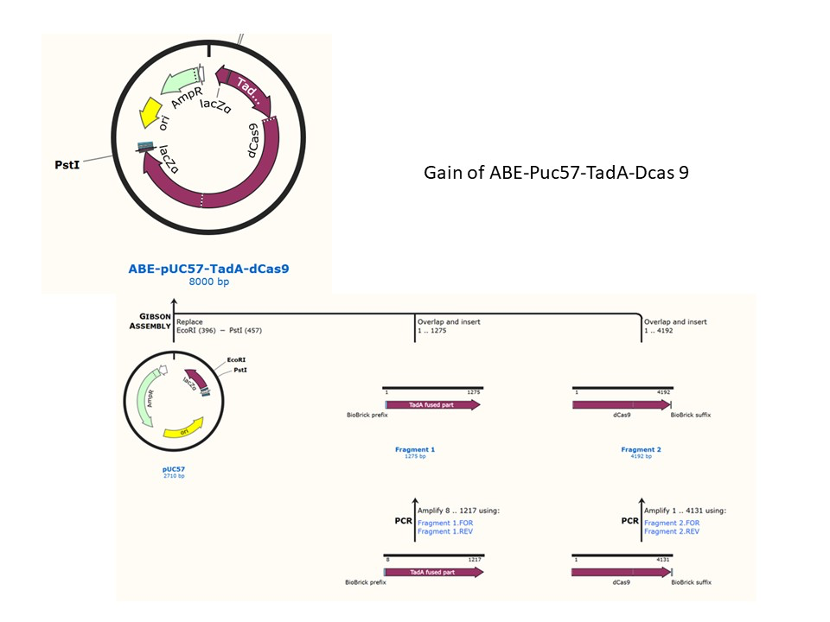
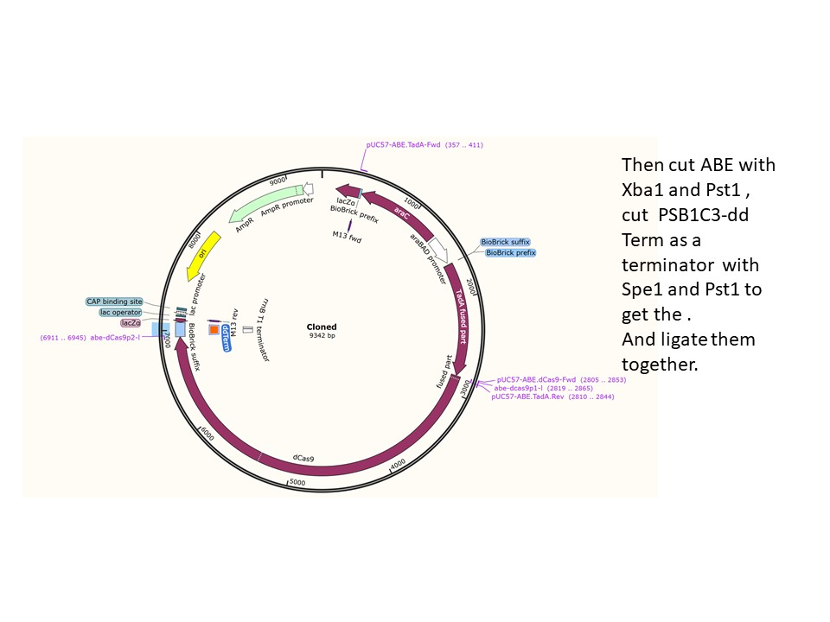
We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as follows:
ABE
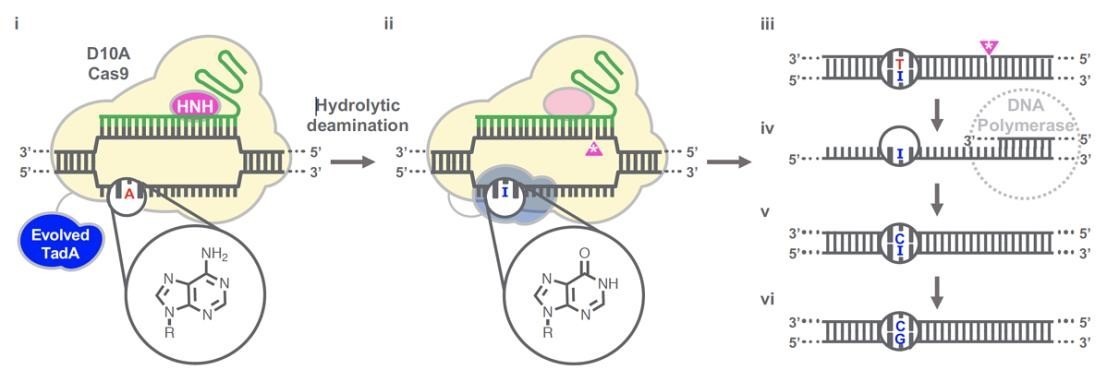
(Gaudelli et al., 2020.)
Gaudelli et al. have successfully developed an adenosine deaminase, which can act on DNA for adenine base editing. They first created a defective chloramphenicol resistance gene (CamR) by introducing a point mutation (H193Y). Reversal of this mutation by adenine base editor will restore antibiotic resistance. To find such a protein, they created a mutant library of E.coli tRNA adenosine deaminase (ecTadA), fused it with dcas9, and transformed it into E.coli containing the defective CamR gene. Screening of viable colonies and subsequent rounds of evolution and engineering produced a mutant TadA (TadA *), which accepted DNA as a substrate satisfactorily.
The artificially evolved adenosine deaminase catalyzes the transformation of target "A" into "I" (inosine), which is regarded as "G" by cell polymerase. Subsequently, a primitive genome A•T base pair was transformed into a G•C base pair. Since inosine excision repair is not as active as uracil excision, ABE does not require any additional inhibitor proteins, such as UGI in CBE.