As part of our project, we grafted the CDRs of mAbs (monoclonal antibodies) against the sex hormones estradiol (E2) and progesterone onto the 3DWT scaffold to obtain nanobodies. As CDR acceptor, also called "scaffold", we used the nanobody cAbBCII-10 [1] (or also NbBcII10 [2] or 3DWT [3], from here on called "3DWT"). This nanobody is also available as part of a composite part in the iGEM parts reg (BBa_K2082001). However, the nanobody is not available as basic part, so we contributed to future iGEM teams in submitting the basic part of cAbBCII-10 (BBa_K3410001). We used the sequence deposited in the PDB by Vincke et al. [2], [3]. Originally, 3DWT was isolated as an antibody capturing and neutralizing enzymes of the β-lactamase B class [1]. β-lactamases are enzymes that hydrolyses β-lactam rings, thus mediating resistance to β-lactam antibiotics and are produced by numerous bacteria. β-lactamases can be divided into classes A, C and D due to differences in their amino acid sequence. Furthermore, they are assigned to the class B of metalloenzymes [4]. 3DWT is suitable as a scaffold, both as an acceptor of CDRs of antibodies from the same taxonomy [5], [6] as well as from other taxonomies [7]. Furthermore, 3DWT has been humanized, meaning that 3DWT is suitable for the use, for example, for human treatment without causing an immune reaction [2].

Figure 1: Structure of the nanobody 3DWT. Colored in red are its CDRs and in grey are the framework regions depicted. This map was created with SnapGene.
The amino acid sequence of 3DWT is as follows:
QVQLVESGGGSVQAGGSLRLSCTASGGSEYSYSTFSLGWFRQAPGQEREAVAAIASMGGLTYYAD
SVKGRFTISRDNAKNTVTLQMNNLKPEDTAIYYCAAVRGYFMRLPSSHNFRYWGQGTQVTVSS
The successful grafting of a new nanobody requires to find an antibody against your antigen of interest (AOI) with an openly available sequence, in order to use its complementarity-determining regions (CDRs), positioned on its variable heavy chain (VH). In a first step of in silico nanobody grafting, the amino acid sequences of both donor (e.g. your antibody against your AOI) and acceptor (e.g. 3DWT) are numbered. There are different numbering schemes for immunoglobulin variable domains available [8]. We decided to use the AHo numbering scheme, because placement of gaps is based on the spatial alignment of known three dimensional structures of immunoglobulin domains and not on sequence variability [9]. The numbering of the VH can be performed using online available tools, we used the tool ANARCI (SAbPred) [10]. For further work, the numbered residues were transferred to an excel sheet. For the grafting process, it is important to consider a series of structurally important framework residues next to the CDRs: These core framework residues have to be grafted from the donor to the acceptor as well [11], [12]. Both, the CDRs and the functional important framework residues of the donor were grafted onto the sequence of the acceptor (3DWT) and a new nanobody is created [7].
Here you can see how the in silico graft looked like in our project for the estradiol nanobody:

Figure 2: Depiction of the grafted estradiol nanobody. The CDRs 1 to 3 (brown) were taken from the estradiol mAb 10G6D6 and grafted onto the scaffold 3DWT (grey). For cloning purposes, SfiI restriction sites were added at 5´ and 3´-ends. Furthermore, to immobilize the nanobody after affinity maturation onto the gold surface of our chip, a his tag was added at the 3´-end. This map was created with SnapGene.
Table 1: Grafting table of the estradiol nanobody based on the sequences of 10G6D6 and 3DWT. Highlighted in yellow are the CDRs 1 to 3 according to AHo. Numbering of the amino acids was done with the AHo numbering scheme.
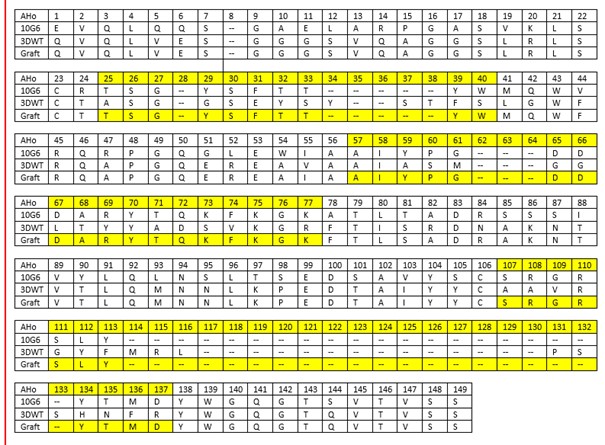
We used 3DWT as a scaffold for our grafting approach for the generation of new and synthetic nanobodies against progesterone and estradiol. We chose the same approach for both hormones with an identical workflow. Shown here are the results for our estradiol nanobody.
Using ChimeraX [13], we optically verified the 3D structure of the grafted nanobodies for steric obstacles to antigen binding: For example, we checked, if the grafted residues sterically impaired residues from the framework:

Figure 3: Screenshot taken from ChimeraX analysing the estradiol graft. An alignment between the scaffold 3DWT and CDR donor (10G6D6) was performed to check for sterical impairments with the amino acid residues. Depicted in yellow is estradiol. Highlighted in cyan is the scaffold 3DWT and in grey 10G6D6. Furthermore, shown in red are the three to be grafted CDRs from 10G6D6. Searching for impairments, we checked every amino acid residue for sterical clashes between the grafted amino acids and the remaining residues from the scaffold which are not neutralized by the grafting process.
Cloning and transformation of the initial grafts into the E. coli strain ER2738 was tested by colony PCR and did work. In a next step, the via epPCR generated library will be cloned and transformed into ER2738.

Figure 4: Colony PCR of ER2738 clones after one day of incubation at 37 °C. The one colony tested, was positive for the estradiol nanobody insert. The gel (1,2% agarose with 1x TAE) was run at 100 V for 20 minutes. A 100 bp ladder was used for size comparison.
By grafting the CDRs from a monoclonal antibody against Estradiol or Progesterone onto the CDRs from 3DWT, a new and synthetic nanobody is generated. But not only the CDRs are grafted, some for the stability important core framework residues from the donor antibody´s VH chain are grafted too. Through affinity maturation measures like Phage Display Technology, a strong binder can be established. In our project we could not obtain strong binding nanobodies with our initial grafted nanobodies, which was to be expected. Consequently, their binding affinity and specifity has to be improved using suitable affinity maturation methods like phage display [7].
We have confirmed that 3DWT can be used as scaffold for a grafting approach and provided its amino acid sequence as a basic part for future iGEM generations.
