Difference between revisions of "Part:BBa K3580101:Experience"
Yuya Otsuki (Talk | contribs) (→Applications of BBa_K3580101) |
Yuya Otsuki (Talk | contribs) (→2.2.1 Cell growth and extract preparation) |
||
| (22 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
===Applications of BBa_K3580101=== | ===Applications of BBa_K3580101=== | ||
In our project, we inserted this parts into pBbe5a(AmpR colE1-ori high copy plasmid., BglBrick vectors) plasmid. | In our project, we inserted this parts into pBbe5a(AmpR colE1-ori high copy plasmid., BglBrick vectors) plasmid. | ||
| + | ==1. <i>in vivo</i> monoterpene synthesis== | ||
| + | ===1.1 Introduction=== | ||
| + | [[File:T--Waseda--monoterpene_3-3-2-1_Schematic_of_in_vivo_limonene_synthesis.png|1500px|thumb|center|Fig. 1-1 Schematic of <i>in vivo</i> monoterpene synthesis]] | ||
| + | Before monoterpene synthesis with cell-free system, we confirmed activity of the genes by monoterpene production with <i>E.coli</i> In this experiment, DH5-alpha transformed with a plasmid encoding enzymes of the mevalonate pathway (pBbA5c-MevT-MBI from addgene) and a plasmid encoding GPP synthetase and monoterpene synthase ([https://parts.igem.org/Part:BBa_K3580101 BBa_K3580101] / [https://parts.igem.org/Part:BBa_K3580102 BBa_K3580102]) were used. | ||
| − | ==<i>in vivo</i> limonene synthesis== | + | ===1.2 Materials and Methods=== |
| + | |||
| + | ===1.2.1 Growth condition=== | ||
| + | DH5-alpha transformed with pBbA5c-MevT-MBI and BBa_K3580101 / BBa_K3580102 were used in this experiment. DH5-alpha was selected as the strain used because it was based on the strain used in the 2019 XJTU-CHINA team's limonene production project. And DH5-alpha transformed with only pBbA5c-MevT-MBI was used as negative control. Strains of transformed DH5-alpha were grown in 2xYT media (16 g/L tryptone, 10 g/L yeast extract, 7 g/L NaCl. These components were purchased from Nakalai tesque) containing 25 ng/µl (Nakalai tesque) of chloramphenicol and 1 % in mass percent concentration of glucose (Wako). After culturing overnight (37℃, 180 rpm, 14 h) on 3 ml scales, 500 µM IPTG (Nacalai tesque) to induce expression and 500 µl dodecane overlay to capture monoterpene were added when OD600 exceeded 1.5. After induction by IPTG, strains were continuously cultured at 30 ℃ and 200 rpm for 2 days (48 h). After 2 days of culture, 300 µl dodecane overlays were collected, mixed with 700 µl ethyl acetate and analyzed by GC/MS. | ||
| + | |||
| + | ===1.2.2 Method of GC/MS analysis=== | ||
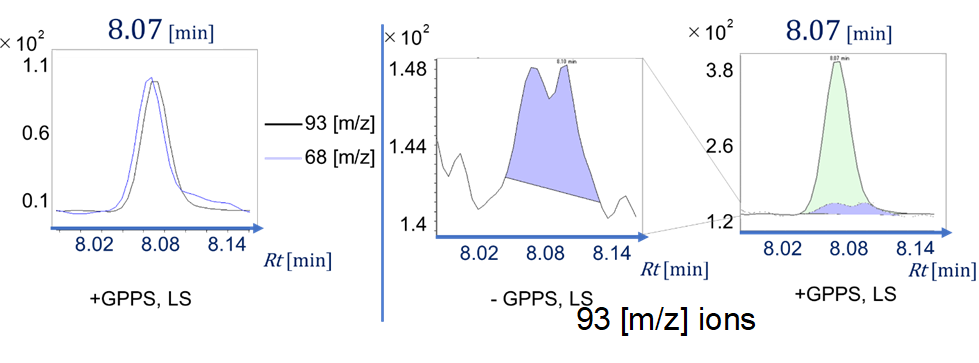
| + | GC/MS analysis was performed on an Agilent 7890B GC with Agilent 5977B MS using a constant flow rate of 1.2 ml/min of helium carrier gas and using HP-5MS (30 m, 0.25 mm, 0.25 µm) column. The inlet temperature was 300 ℃ and initial column temperature was 40 ℃ for 1 min. The column was then heated to 300 ℃ at a rate of 15 ℃/min and held at 300 ℃ for 2 min. The Injection volume was 1 µl with the split ratio 10:1. In this experiment, SIM mode was used for the analysis. Ions with four m/z values characteristic of limonene (68 and 93) and sabinene (77 and 91, 93) were analyzed to confirm the presence of monoterpenes. And for quantitative analysis, 93 m/z was used in each monoterpene analyzes. The quantification of monoterpenes was performed by comparing the analysis results with the data of each monoterpene 100 µM standard product. These m/z values were selected by referring to the mass spectrum of each monoterpene present in NIST Standard Reference database. Preliminary experiments have confirmed that the retention time for the peaks of each monoterpene is 8.07 min for limonene and 8.45 min for sabinene (Fig. 1-2). | ||
| + | [[File:T--Waseda--monoterpene_3-3-1-1_Major_peak_for_standard_products_of_each_monoterpene.png|1500px|thumb|center|Fig. 1-2 Major peak for standard products of each monoterpene]] | ||
| + | |||
| + | ===1.3 Results=== | ||
| + | From GC/MS analysis, we confirmed limonene from the introduced pathway. Ions with two characteristic m/z values (68, 93) in limonene were detected in the limonene synthesis system using BBa_K3580101. Also, GC chromatogram of the ion having 93 m/z from the complete limonene pathway shows a peak higher than that from a negative control (Fig. 1-3). Therefore, the yield of synthesized limonene per culture solution was calculated to be 0.161 µM. | ||
| + | [[File:T--Waseda--monoterpene_3-3-2-2_GCMS_analysis_results_of_in_vivo_limonene_synthesis_system.png|700px|thumb|center|Fig. 1-3 GC/MS analysis results of <i>in vivo</i> limonene synthesis system]] | ||
| + | |||
| + | ==2. Cell-free monoterpene synthesis== | ||
| + | ===2.1 Introduction=== | ||
| + | [[File:T--waseda--monoterpene_2-2-1_parts_and_metabolic_pathways_in_this_experiment_and_a_schematic_diagram_of_the_experiment.png|1500px|thumb|center|Fig. 2-1 Parts and metabolic pathways in the experiment of monoterpene synthesis and a schematic diagram of the experiment ]] | ||
| + | |||
| + | In this cell-free monoterpene synthesis, we mixed two E. coli extracts each of which has either first 7 or last 2 enzymes of a pathway from Ac-CoA, which is a major intermediate of cell central metabolism. Through mevalonate pathway, the former extract one (derived from E. coli into which pBbA5c-MevT-MBI has been introduced) can provide IPP and DMAPP, which can also be used as intermediates for other important biosynthesis. Here we indeed supplemented only glucose and acetate as carbon sources. We obtained expression system for those seven genes from addgene and have converted this into Biobrick RFC 1000 format by synonymous replacement ([https://parts.igem.org/Part:BBa_K3580103 BBa_K3580103]). In order to take advantage of an engineering principle of synthetic biology we provided two biobrick parts ( [https://parts.igem.org/Part:BBa_K3580101 BBa_K3580101], [https://parts.igem.org/Part:BBa_K3580102 BBa_K3580102]) for the source for the latter extract. BBa_K3580101 has GPP synthase (GPPS) and limonene synthase. Although GPP synthase is shared with BBa_K3580101, BBa_K3580102 has sabinene synthase, which has one point mutation in limonene synthase (Srividya Narayanan et al 2015) and a new coding sequence for Parts registry of iGEM (See here for more details on this experiments). | ||
| + | |||
| + | ===2.2 Materials and Methods=== | ||
| + | ===2.2.1 Cell growth and extract preparation=== | ||
| + | Transformed DH5-α strains were grown in 2xYT medium containing 25 ng / µl chloramphenicol and 1% glucose concentration. This is the same as in vivo monoterpene synthesis. After culturing overnight (37℃, 180 rpm, 14 h) on 6 ml scales, 6 ml culture were added to 100 ml 2xYT medium containing 25 ng / µl chloramphenicol and 1% glucose concentration. Then, the culture was continued at 37 ℃ and 250 rpm, and when OD600 exceeded 0.6, 500 µM IPTG and 500 µL dodecane overlay were added. After that, the strains into which BBa_K3580102 (Ptrc-trGPPS-SS) or pBbA5c-MevT-MBI was introduced were continuously cultured for 30℃, 250rpm, 22 h, and the strains into which BBa_K3580101 (Ptrc-trGPPS-LS) was introduced were continuously cultured for 30℃, 250rpm, 6 h. [https://2020.igem.org/Team:Waseda/Protocol After culturing, a cell extract was prepared according to step 3 and subsequent steps of the extract preparation protocol]. However, the Buffer E used did not contain dithiothreitol. | ||
| + | |||
| + | ===2.2.2 Preparation of reaction solution for monoterpene synthesis and reaction conditions=== | ||
| + | The preparation of the reaction solution for monoterpene synthesis is based on a recent paper from Jwett lab (Dudly et al 2019). 100 µl fraction of the mixed extract and 100 µl fraction of the solution containing the substrate etc. were mixed to prepare the 200 µl reaction solution. The reaction contains 200 mM glucose, 8 mM magnesium acetate (Nakalai tesque), 10 mM ammonium acetate (Nakalai tesque), 134 mM potassium acetate (Nakalai tesque), 100 mM Bis Tris buffer (Bis Tris was purchased from Sigma, hydrochloric acid was purchased from Nakalai tesque), 10 mM potassium phosphate (Nakalai tesque), and the enzymes contained in the extract. The ratio of enzymes contained in the reaction solution could be adjusted by changing the mixing ratio of the extracts. 200 µl dodecane overlay was added to the prepared reaction solutions, and the reaction solutions were incubated at 30 ℃ for 2 days. After two days reaction, 150 µl dodecane overlay was collected, mixed with 150 µl dodecane and 700 µl ethyl acetate, and then analyzed by GC/MS. The GC / MS methods used were as described in 1.2.2. | ||
| + | |||
| + | ===2.3 Results and Discussion=== | ||
| + | [[File:T--Waseda--monoterpene_2-2-2_GCMS_analysis_results_of_limonene_synthesis_system.png|1500px|thumb|center|Fig. 2-2 GC/MS analysis results of cell-free limonene synthesis system]] | ||
| + | |||
| + | We confirmed limonene synthesis GC/MS analysis with SIM. In this SIM analysis, ions with four m/z values characteristic in limonene (68 and 93) and sabinene (77 and 91, 93) were analyzed. By using authentic limonene standard, we confirmed a retention time for GC and the characteristic limonene SIM signal at the specific m/z values. At the same retention time with the standard, limonene-specific m/z value (68, 93) ions were detected in the selected ions (The upper right figure of Fig. 2-2). We also draw a GC chart by summation of the signals from the selected ions (The lower left figure of Fig. 2-2). By comparison with a negative control experiment which we omitted the extract containing GPP synthase and limonene synthase, we found clear peak from our limonene synthesis. | ||
| + | |||
| + | [[File:T--Waseda--monoterpene_2-2-4_Yields_of_monoterpene_per_reaction_solution_in_this_experiment.png|1500px|thumb|center|Fig. 2-3 Yields of monoterpene per reaction solution in this experiment]] | ||
| + | |||
| + | Finally, each of monoterpenes were quantified based on peaks of substance having 93 m/z and each monoterpene standard curves. Taking advantage of the modularity of the combination of extracts, we confirmed whether the yield of monoterpenes could be changed by changing the mixing ratio of the extract containing the enzyme of the mevalonate pathway and the extract containing GPP synthase and monoterpene synthase. As a result, changes in the yield of monoterpenes due to the mixing ratio of the extracts were observed (Fig. 2-3). The best yield of limonene per reaction solution with limonene synthase contained system was 0.73 µM. | ||
| + | |||
| + | ==Reference== | ||
| + | (1) Alonso-Gutierrez, J. et al. (2013). Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic engineering, 19, 33-41. | ||
| + | |||
| + | (2) Dudley, Q. M. et al. (2019). Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synthetic Biology, 4(1), ysz003. | ||
| + | |||
| + | (3) Srividya, N. et al. (2015). Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proceedings of the National Academy of Sciences, 112(11), 3332-3337. | ||
| + | |||
| + | (4) Korman, T. P. et al. (2017). A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nature communications, 8(1), 1-8. | ||
| + | |||
| + | (5) Mass Spectrometry Data Center, William E. Wallace, "Mass Spectra" in NIST Chemistry WebBook, NIST Standard Reference Database Number 69, Eds. P.J. Linstrom and W.G. Mallard, National Institute of Standards and Technology, Gaithersburg MD, 20899, https://doi.org/10.18434/T4D303, (retrieved October 22, 2020). | ||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 00:44, 3 December 2020
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K3580101
In our project, we inserted this parts into pBbe5a(AmpR colE1-ori high copy plasmid., BglBrick vectors) plasmid.
1. in vivo monoterpene synthesis
1.1 Introduction
Before monoterpene synthesis with cell-free system, we confirmed activity of the genes by monoterpene production with E.coli In this experiment, DH5-alpha transformed with a plasmid encoding enzymes of the mevalonate pathway (pBbA5c-MevT-MBI from addgene) and a plasmid encoding GPP synthetase and monoterpene synthase (BBa_K3580101 / BBa_K3580102) were used.
1.2 Materials and Methods
1.2.1 Growth condition
DH5-alpha transformed with pBbA5c-MevT-MBI and BBa_K3580101 / BBa_K3580102 were used in this experiment. DH5-alpha was selected as the strain used because it was based on the strain used in the 2019 XJTU-CHINA team's limonene production project. And DH5-alpha transformed with only pBbA5c-MevT-MBI was used as negative control. Strains of transformed DH5-alpha were grown in 2xYT media (16 g/L tryptone, 10 g/L yeast extract, 7 g/L NaCl. These components were purchased from Nakalai tesque) containing 25 ng/µl (Nakalai tesque) of chloramphenicol and 1 % in mass percent concentration of glucose (Wako). After culturing overnight (37℃, 180 rpm, 14 h) on 3 ml scales, 500 µM IPTG (Nacalai tesque) to induce expression and 500 µl dodecane overlay to capture monoterpene were added when OD600 exceeded 1.5. After induction by IPTG, strains were continuously cultured at 30 ℃ and 200 rpm for 2 days (48 h). After 2 days of culture, 300 µl dodecane overlays were collected, mixed with 700 µl ethyl acetate and analyzed by GC/MS.
1.2.2 Method of GC/MS analysis
GC/MS analysis was performed on an Agilent 7890B GC with Agilent 5977B MS using a constant flow rate of 1.2 ml/min of helium carrier gas and using HP-5MS (30 m, 0.25 mm, 0.25 µm) column. The inlet temperature was 300 ℃ and initial column temperature was 40 ℃ for 1 min. The column was then heated to 300 ℃ at a rate of 15 ℃/min and held at 300 ℃ for 2 min. The Injection volume was 1 µl with the split ratio 10:1. In this experiment, SIM mode was used for the analysis. Ions with four m/z values characteristic of limonene (68 and 93) and sabinene (77 and 91, 93) were analyzed to confirm the presence of monoterpenes. And for quantitative analysis, 93 m/z was used in each monoterpene analyzes. The quantification of monoterpenes was performed by comparing the analysis results with the data of each monoterpene 100 µM standard product. These m/z values were selected by referring to the mass spectrum of each monoterpene present in NIST Standard Reference database. Preliminary experiments have confirmed that the retention time for the peaks of each monoterpene is 8.07 min for limonene and 8.45 min for sabinene (Fig. 1-2).
1.3 Results
From GC/MS analysis, we confirmed limonene from the introduced pathway. Ions with two characteristic m/z values (68, 93) in limonene were detected in the limonene synthesis system using BBa_K3580101. Also, GC chromatogram of the ion having 93 m/z from the complete limonene pathway shows a peak higher than that from a negative control (Fig. 1-3). Therefore, the yield of synthesized limonene per culture solution was calculated to be 0.161 µM.
2. Cell-free monoterpene synthesis
2.1 Introduction
In this cell-free monoterpene synthesis, we mixed two E. coli extracts each of which has either first 7 or last 2 enzymes of a pathway from Ac-CoA, which is a major intermediate of cell central metabolism. Through mevalonate pathway, the former extract one (derived from E. coli into which pBbA5c-MevT-MBI has been introduced) can provide IPP and DMAPP, which can also be used as intermediates for other important biosynthesis. Here we indeed supplemented only glucose and acetate as carbon sources. We obtained expression system for those seven genes from addgene and have converted this into Biobrick RFC 1000 format by synonymous replacement (BBa_K3580103). In order to take advantage of an engineering principle of synthetic biology we provided two biobrick parts ( BBa_K3580101, BBa_K3580102) for the source for the latter extract. BBa_K3580101 has GPP synthase (GPPS) and limonene synthase. Although GPP synthase is shared with BBa_K3580101, BBa_K3580102 has sabinene synthase, which has one point mutation in limonene synthase (Srividya Narayanan et al 2015) and a new coding sequence for Parts registry of iGEM (See here for more details on this experiments).
2.2 Materials and Methods
2.2.1 Cell growth and extract preparation
Transformed DH5-α strains were grown in 2xYT medium containing 25 ng / µl chloramphenicol and 1% glucose concentration. This is the same as in vivo monoterpene synthesis. After culturing overnight (37℃, 180 rpm, 14 h) on 6 ml scales, 6 ml culture were added to 100 ml 2xYT medium containing 25 ng / µl chloramphenicol and 1% glucose concentration. Then, the culture was continued at 37 ℃ and 250 rpm, and when OD600 exceeded 0.6, 500 µM IPTG and 500 µL dodecane overlay were added. After that, the strains into which BBa_K3580102 (Ptrc-trGPPS-SS) or pBbA5c-MevT-MBI was introduced were continuously cultured for 30℃, 250rpm, 22 h, and the strains into which BBa_K3580101 (Ptrc-trGPPS-LS) was introduced were continuously cultured for 30℃, 250rpm, 6 h. After culturing, a cell extract was prepared according to step 3 and subsequent steps of the extract preparation protocol. However, the Buffer E used did not contain dithiothreitol.
2.2.2 Preparation of reaction solution for monoterpene synthesis and reaction conditions
The preparation of the reaction solution for monoterpene synthesis is based on a recent paper from Jwett lab (Dudly et al 2019). 100 µl fraction of the mixed extract and 100 µl fraction of the solution containing the substrate etc. were mixed to prepare the 200 µl reaction solution. The reaction contains 200 mM glucose, 8 mM magnesium acetate (Nakalai tesque), 10 mM ammonium acetate (Nakalai tesque), 134 mM potassium acetate (Nakalai tesque), 100 mM Bis Tris buffer (Bis Tris was purchased from Sigma, hydrochloric acid was purchased from Nakalai tesque), 10 mM potassium phosphate (Nakalai tesque), and the enzymes contained in the extract. The ratio of enzymes contained in the reaction solution could be adjusted by changing the mixing ratio of the extracts. 200 µl dodecane overlay was added to the prepared reaction solutions, and the reaction solutions were incubated at 30 ℃ for 2 days. After two days reaction, 150 µl dodecane overlay was collected, mixed with 150 µl dodecane and 700 µl ethyl acetate, and then analyzed by GC/MS. The GC / MS methods used were as described in 1.2.2.
2.3 Results and Discussion
We confirmed limonene synthesis GC/MS analysis with SIM. In this SIM analysis, ions with four m/z values characteristic in limonene (68 and 93) and sabinene (77 and 91, 93) were analyzed. By using authentic limonene standard, we confirmed a retention time for GC and the characteristic limonene SIM signal at the specific m/z values. At the same retention time with the standard, limonene-specific m/z value (68, 93) ions were detected in the selected ions (The upper right figure of Fig. 2-2). We also draw a GC chart by summation of the signals from the selected ions (The lower left figure of Fig. 2-2). By comparison with a negative control experiment which we omitted the extract containing GPP synthase and limonene synthase, we found clear peak from our limonene synthesis.
Finally, each of monoterpenes were quantified based on peaks of substance having 93 m/z and each monoterpene standard curves. Taking advantage of the modularity of the combination of extracts, we confirmed whether the yield of monoterpenes could be changed by changing the mixing ratio of the extract containing the enzyme of the mevalonate pathway and the extract containing GPP synthase and monoterpene synthase. As a result, changes in the yield of monoterpenes due to the mixing ratio of the extracts were observed (Fig. 2-3). The best yield of limonene per reaction solution with limonene synthase contained system was 0.73 µM.
Reference
(1) Alonso-Gutierrez, J. et al. (2013). Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic engineering, 19, 33-41.
(2) Dudley, Q. M. et al. (2019). Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synthetic Biology, 4(1), ysz003.
(3) Srividya, N. et al. (2015). Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proceedings of the National Academy of Sciences, 112(11), 3332-3337.
(4) Korman, T. P. et al. (2017). A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nature communications, 8(1), 1-8.
(5) Mass Spectrometry Data Center, William E. Wallace, "Mass Spectra" in NIST Chemistry WebBook, NIST Standard Reference Database Number 69, Eds. P.J. Linstrom and W.G. Mallard, National Institute of Standards and Technology, Gaithersburg MD, 20899, https://doi.org/10.18434/T4D303, (retrieved October 22, 2020).
User Reviews
UNIQe9bf8a9ff2618f65-partinfo-00000000-QINU UNIQe9bf8a9ff2618f65-partinfo-00000001-QINU