Difference between revisions of "Part:BBa K274200:Experience"
Juliechiang (Talk | contribs) (→User Reviews) |
Juliechiang (Talk | contribs) (→User Reviews) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 13: | Line 13: | ||
===User Reviews=== | ===User Reviews=== | ||
<!-- DON'T DELETE --><partinfo>BBa_K274200 StartReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K274200 StartReviews</partinfo> | ||
| − | <!-- Template for a user review | + | <!-- Template for a user review --> |
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
| − | |||
<partinfo>BBa_K274200 AddReview number</partinfo> | <partinfo>BBa_K274200 AddReview number</partinfo> | ||
| − | <I> | + | <I>NCTU_Formosa</I> |
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | + | From the characterization performed by Peking iGEM Team 2010, we learn that a promoter is not required to express CrtEBIY. To investigate this interesting discovery further, we utilized the tool: Promoter Prediction by Neural Network (Martin Reese, Lawquenrence Berkeley Laboratory, CA, U.S.A.) - applicable to eukaryotes and prokarotes (Reference: Reese MG, 2001. Comput Chem 26: 51-56). Intriguingly, we discovered many promoter-like sequences in the front of the sequences, shown in Fig. 1. These results provide a possible explanation as to why this part can express CrtEBIY without regulation. | |
| − | + | <html><a href="https://2020.igem.org/wiki/images/2/28/T--NCTU_Formosa--hunter.jpg" target="blank"><img style="width:60%; " src="https://2020.igem.org/wiki/images/2/28/T--NCTU_Formosa--hunter.jpg" ></a></html> | |
| − | < | + | <br><font>Figure 1: Result of promoter prediction</font> |
| − | + | <font>We conducted colony PCR and double confirmed with digestion to verify that BBa_K274200 was correctly cloned into the E. coli Lemo21 (DE3).</font><br> | |
| − | + | <html><a href="https://2020.igem.org/wiki/images/e/ea/T--NCTU_Formosa--exp-result4.png" target="blank"><img style="width:20%; " src="https://2020.igem.org/wiki/images/e/ea/T--NCTU_Formosa--exp-result4.png" ></a></html> | |
| − | + | <html><a href="https://2020.igem.org/wiki/images/3/33/T--NCTU_Formosa--exp-result5.png" target="blank"><img style="width:20%; " src="https://2020.igem.org/wiki/images/3/33/T--NCTU_Formosa--exp-result5.png" ></a></html> | |
| − | < | + | <br><font>Figure 2: Colony PCR & Digest Result of BBa_K274200</font> |
| − | < | + | <font>Carotene Extraction in acetone</font> |
| − | + | <font>We conducted colony PCR and digest to verify that BBa_K274200 was correctly cloned into <i>E. coli</i> Lemo21 (DE3).</font> | |
| − | + | <html><a href="https://2020.igem.org/wiki/images/0/07/T--NCTU_Formosa--expresult6.jpg" target="blank"><img style="width:60%; " src="https://2020.igem.org/wiki/images/0/07/T--NCTU_Formosa--expresult6.jpg" ></a></html> | |
| + | <br><font>Figure 3: β-carotene Extraction Absorption Spectrum</font> | ||
| + | <html><a href="https://2020.igem.org/wiki/images/c/c4/T--NCTU_Formosa--colltas.jpg" target="blank"><img style="width:60%; " src="https://2020.igem.org/wiki/images/c/c4/T--NCTU_Formosa--colltas.jpg" ></a></html> | ||
| + | <br><font>Figure 4: TAS_Taipei Validation(green line): β-carotene extraction absorption spectrum</font> | ||
| + | |} | ||
| − | |||
| − | |||
<!-- DON'T DELETE --><partinfo>BBa_K274200 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K274200 EndReviews</partinfo> | ||
Latest revision as of 16:09, 27 October 2020
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K274200
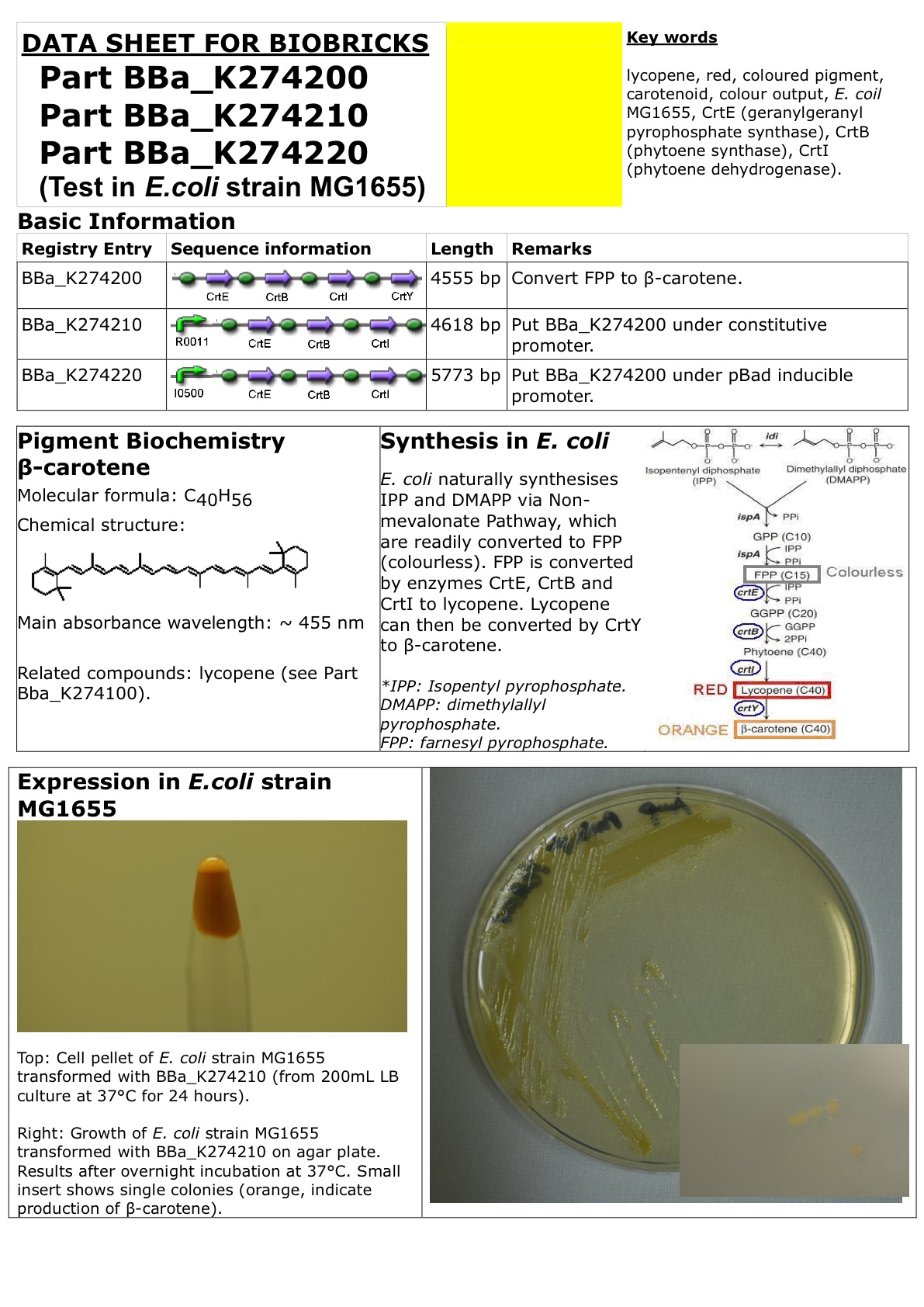
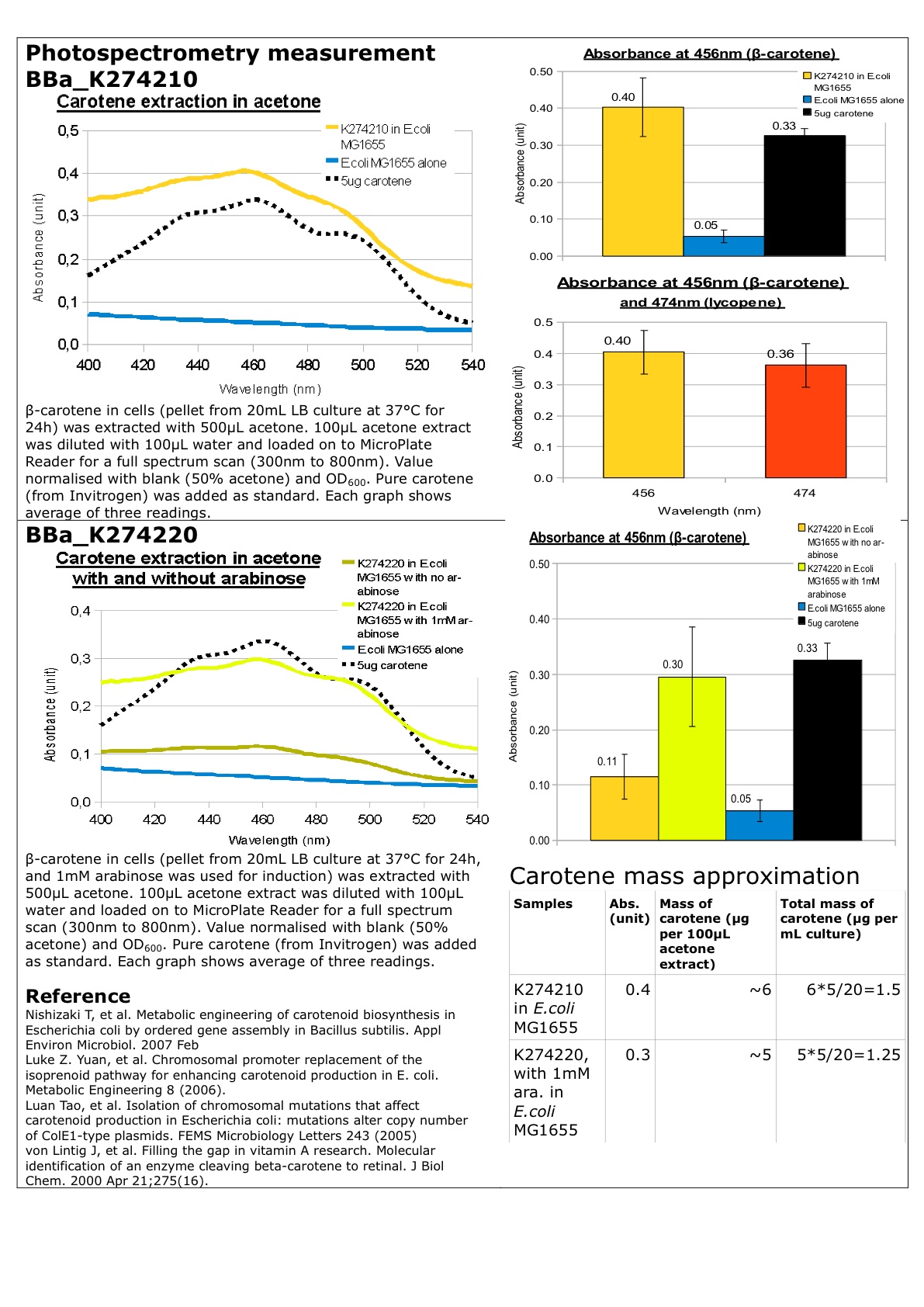
Datasheet for Part BBa_K274200, Part BBa_274210 and Part BBa_274220 in E.coli strain MG1655.
For PDF version of this Datasheet: File:Carotene.pdf
User Reviews
UNIQ97556f6f0459998c-partinfo-00000000-QINU
UNIQ97556f6f0459998c-partinfo-00000007-QINU
Characterization by Peking iGEM Team 2010
We find that BBa_K274200 represented a significant leakage expression when exploited as a reporter gene and even bacteria bearing BBa_K274200 only could also represent significant color change, compared with the blank. We speculate that it’s because of a putative promoter upstream, resulting in the leaky expression of CrtEBIY. In order to verify the speculation, we further characterize this biobrick. This biobrick was suffixed to the constitutive promoter BBa_J23103. Resulted new biobrick is shown in Figure 1.

Figure 1. The biobrick we constructed to characterize biobrick BBa_K274200. It is Constitutive promoter BBa_J23103 driving CrtEBIY
After the biobrick was constructed, it was transformed to JM109, BL21 (DE3) and DH5α, respectively. Then we compared their color with the corresponding strains that bear plasmid with only CrtEBIY as the insert.
After cultivated at 37℃ for 12 hours, as shown in Figure 8, bacteria bearing BBa_J23103-CrtEBIY and naked CrtEBIY represented a yellow color and blank vector appeared white (Fig 2).

Constitutive promoter BBa_J23103-CrtEBIY
Figure 2 Color contrast after 12 hours’ cultivation (for each photo, from the left to the right are JM109, DH5α and BL21)
From these pictures above, we can see that both strains bearing constitutive promoter-CrtEBIY and strains bearing naked CrtEBIY appeared yellow. This demonstrated that CrtEBIY has a serious leakage, and the cause of leakage is probably the same as CrtEBI. It is noteworthy that CrtEBIY pathway need one more enzyme (Lycopene Cyclase, produced by CrtY) than CrtEBI and the Km for Lycopene Cyclase is 1.8μM (Schnurr et al., 1996), so the leakage was more serious as expected.
We intended to use CrtEBI and CrtEBIY as our bioreporter at first. After our characterization, we decided to use CrtEBI for the serious leakage of CrtEBIY. However, we found a serious leakage when CrtEBI was combined with the promoter PmerT and PpbrA. Leakage was serious regardless of the prefixed promoter in our experiment. For this reason, we chose other reporter gene, such as LacZ alpha instead of CrtEBI.
Characterisation by Wits South Africa 2010
Our Lacto-Report construct design included an E.chromi Biobrick BBa_k274200.
However, after a number of attempts to assemble this part into our machine using the Standard Assembly method with no success, we decided to determine if all the correct Biobrick restriction sites were present. We also suspected that perhaps one of the sites had been methylated, so we transformed the initial Biobrick plasmid received from the DNA distribution into a methylase negative strain of E.coli (Sure cells). We then prepared plasmid from one of the colonies and performed a series of digests on the plasmid.
Figure 1. E.chromi Orange digested with various restriction enzymes to confirm the presence of the Biobrick restriction sites.
In Figure 1, the plasmid sample digested with Spe1 (Fermentas) resembles the undigested control. The plasmid sample double-digested with EcoR1 and Spe1 (Fermentas) is linearised by cutting with EcoR1 but there are no distinct insert and plasmid bands, which is the banding pattern we expect to see after digesting with these two enzymes. The expected pattern is clearly visible in the mCherry control which was also digested with EcoR1 and Spe1 (Fermentas).
Thus, we concluded that the Spe1 restriction site was absent in the Orange E.chromi Biobrick.