Difference between revisions of "Part:BBa K165080"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K165080 short</partinfo> | <partinfo>BBa_K165080 short</partinfo> | ||
| + | |||
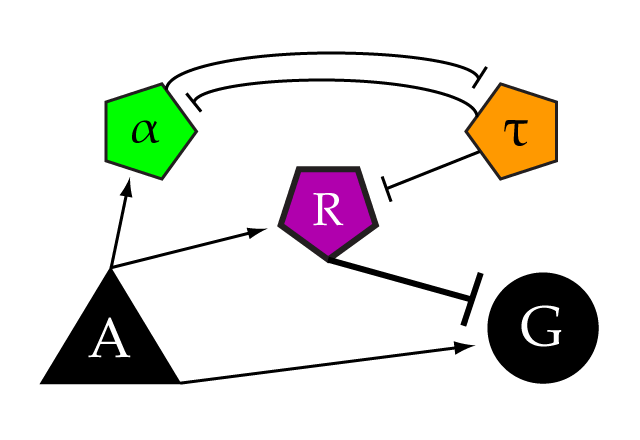
| + | This is the LexA untagged activator driven by pGAL1. It is used as the A element in a limiter construct, modeling an endogenous transcriptional activator for our proof-of-concept. The pGAL promoter normally has an all-or-nothing response. To make it tunable, knock out the Gal2 gene. | ||
| + | |||
| + | |||
| + | [[Image:Glyph_labeled2.png|center|400px]] | ||
| + | |||
| + | |||
| + | |||
| + | ===Usage and Biology=== | ||
| + | |||
| + | To build a (heretofore untested) limiter, this part should be in a yeast strain with the following constructs: | ||
| + | |||
| + | '''A)''' [[Part:BBa_K165080]] pGAL1 + Untagged LexA activator on pRS305<br> | ||
| + | '''G)''' [[Part:BBa_K165078]] LexA activable reporter on pRS306<br> | ||
| + | Into mating type a. Knock out the Gal2 gene for a tunable level of induction.<br> | ||
| + | |||
| + | '''R)''' [[Part:BBa_K165090]] Gli1 bs + LexA bs + mCYC + LexA repressor (mCherryx2 tagged) on pRS306<br> | ||
| + | '''Alpha)''' [[Part:BBa_K165095]] Gli1 bs + LexA bs + mCYC + Zif268-HIV repressor (mCherryx2 tagged) on pRS303 <br> | ||
| + | '''Tau)''' [[Part:BBa_K165096]] Zif268-HIV bs + MET25+ Gli1 repressor (CFPx2 tagged) on pRS304* <br> | ||
| + | Into mating type alpha. Knock out the Gal2 gene for a tunable level of induction.<br> | ||
| + | |||
| + | Mate the two types, select on SD-His-Trp-Leu-Ura | ||
| + | |||
| + | '''Inputs:''' | ||
| + | Set the threshold by tuning [Met] between 0 and 500 uM | ||
| + | Set the level of induction (A) with Galactose between 0-3% | ||
| + | |||
| + | '''Outputs expected:'''<br> | ||
| + | ''Subthreshold [A]:'' CFP in the nucleus from Tau, YFP in the cytosol proportionate to subthreshold induction from G being activated by A and not repressed by R.<br> | ||
| + | ''Superthreshold [A]:'' mCherry in the nucleus from Alpha and R induction. YFP in cytosol levels out at threshold limit despite rising induction. | ||
- | - | ||
Latest revision as of 03:50, 31 October 2008
pGAL1 + Untagged LexA activator on pRS305
This is the LexA untagged activator driven by pGAL1. It is used as the A element in a limiter construct, modeling an endogenous transcriptional activator for our proof-of-concept. The pGAL promoter normally has an all-or-nothing response. To make it tunable, knock out the Gal2 gene.
Usage and Biology
To build a (heretofore untested) limiter, this part should be in a yeast strain with the following constructs:
A) Part:BBa_K165080 pGAL1 + Untagged LexA activator on pRS305
G) Part:BBa_K165078 LexA activable reporter on pRS306
Into mating type a. Knock out the Gal2 gene for a tunable level of induction.
R) Part:BBa_K165090 Gli1 bs + LexA bs + mCYC + LexA repressor (mCherryx2 tagged) on pRS306
Alpha) Part:BBa_K165095 Gli1 bs + LexA bs + mCYC + Zif268-HIV repressor (mCherryx2 tagged) on pRS303
Tau) Part:BBa_K165096 Zif268-HIV bs + MET25+ Gli1 repressor (CFPx2 tagged) on pRS304*
Into mating type alpha. Knock out the Gal2 gene for a tunable level of induction.
Mate the two types, select on SD-His-Trp-Leu-Ura
Inputs: Set the threshold by tuning [Met] between 0 and 500 uM Set the level of induction (A) with Galactose between 0-3%
Outputs expected:
Subthreshold [A]: CFP in the nucleus from Tau, YFP in the cytosol proportionate to subthreshold induction from G being activated by A and not repressed by R.
Superthreshold [A]: mCherry in the nucleus from Alpha and R induction. YFP in cytosol levels out at threshold limit despite rising induction.
-
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 150
- 1000COMPATIBLE WITH RFC[1000]