Difference between revisions of "Part:BBa K3332009"
AnnaTaylor (Talk | contribs) |
AnnaTaylor (Talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3332009 short</partinfo> | <partinfo>BBa_K3332009 short</partinfo> | ||
| − | We anchor GOX protein onto membranes through AIDA to catalyze the reaction of degradating glyphosate to form glyoxalic acid and AMPA. We use K880005 to construct the expression system and anchor GOX on the surface of E.coli. | + | We anchor GOX protein onto membranes through AIDA to catalyze the reaction of degradating glyphosate to form glyoxalic acid and AMPA. We use K880005 to construct the expression system and anchor GOX on the surface of ''E.coli''. |
| Line 12: | Line 12: | ||
<html> | <html> | ||
<figure> | <figure> | ||
| − | <img src="https://2020.igem.org/wiki/images/f/f1/T--XMU-China--XMU-China_2020-Mechanism_of_GOX_and_GRHPR.png" width=" | + | <img src="https://2020.igem.org/wiki/images/f/f1/T--XMU-China--XMU-China_2020-Mechanism_of_GOX_and_GRHPR.png" width="80%" style="float:center"> |
<figcaption> | <figcaption> | ||
<p style="font-size:1rem"> | <p style="font-size:1rem"> | ||
| Line 28: | Line 28: | ||
<html> | <html> | ||
<figure> | <figure> | ||
| − | <img src="https://2020.igem.org/wiki/images/9/9a/T--XMU-China--XMU-China_2020-J23100_B0034_gox-aidA_B0015.png" width=" | + | <img src="https://2020.igem.org/wiki/images/9/9a/T--XMU-China--XMU-China_2020-J23100_B0034_gox-aidA_B0015.png" width="35%" style="float:center"> |
<figcaption> | <figcaption> | ||
<p style="font-size:1rem"> | <p style="font-size:1rem"> | ||
Latest revision as of 22:54, 27 October 2020
GOX-AIDA
We anchor GOX protein onto membranes through AIDA to catalyze the reaction of degradating glyphosate to form glyoxalic acid and AMPA. We use K880005 to construct the expression system and anchor GOX on the surface of E.coli.
Biology
AIDA is an anchor protein from E. coli, which has been widely used in cell-surface display. GOX, also known as EcAKR4-1, is found in Echinochloa colona. It can decompose glyphosate into AMPA and glyoxylic acid. GOX is fused at C terminal with AIDA to get GOX-AIDA and we hope GOX can be displayed on the surface of E. coli.[1][2]
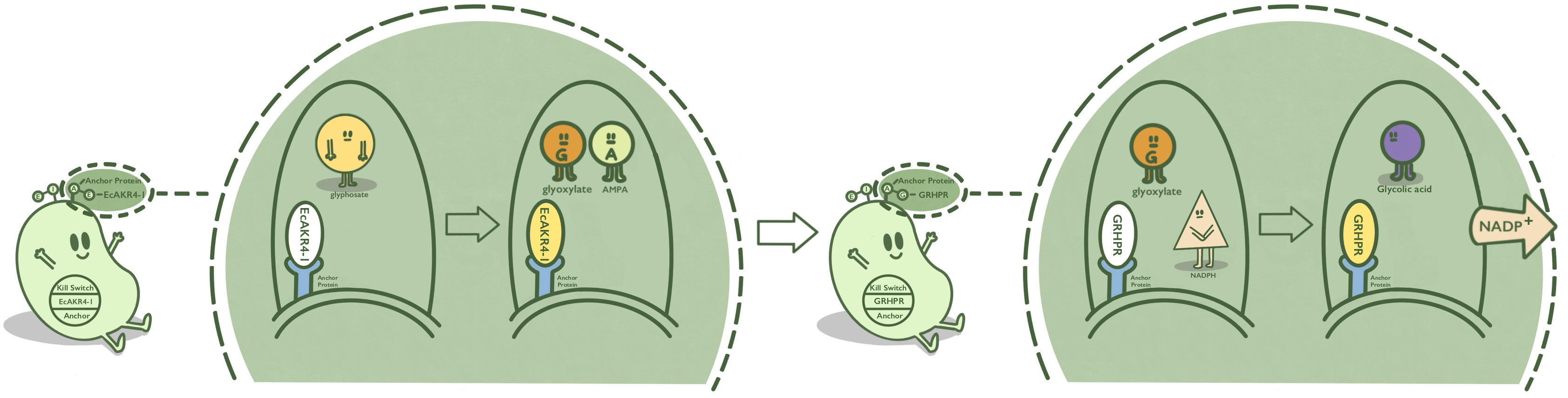
- Fig 1. Mechanism of GOX-AIDA on the surface of E. coli
Usage
Here, we used BBa_K880005 to construct the expression system and obtained the composite part BBa_K3332054. We transformed the constructed plasmid into E. coli BL21 (DE3) to verify its successful expression.

- Fig 2. Gene circuit of GOX-AIDA.
Characterization
1. Identification
After receiving the synthesized DNA, restriction digestion was done to certify that the plasmid was correct, and the experimental results were shown in figure3.

- Fig 3. DNA gel electrophoresis of restriction-digest products of GOX-AIDA-pSB1C3 (Xba I & Pst I sites).
2. Ability of degrading NADPH
After transforming glyphosate into glyoxylic acid with the function of GOX, we use GRHPR, a Glyoxylate reductase from human liver, to reduce glyoxylic acid. GRHPR can convert glyoxylic acid when NADPH is consumed as cofactor. NADPH is a suitable target compound that can be detected by the signal of OD340. And when NADPH is consumed, OD340 declines.
We mixed glyphosate solution, NADPH solution, purified GRHPR protein dissolved in Tris-HCl(pH=7.5) and bacteria solution carrying GOX-AIDA. Then, we immediately detected OD340 by using TECAN® Infinite M200 Pro to see the effect of GOX fused with anchor protein.
When using bacteria carrying GOX-AIDA, we successfully found OD340 decrease as time went on. When using bacteria carrying J23100-RBS(BBa_K880005) and bacteria carrying GOX-Histag as control , we can see the decrease is less than the GOX-AIDA samples. The results prove that GOX-AIDA can be displayed on the surface and convert glyphosate as normal, which is shown in figure 5.

- Fig 4. OD340-Time curve of three fusion protein of GOX and anchor protein
References
- ↑ Pan L, Yu Q, Han H, et al. Aldo-keto Reductase Metabolizes Glyphosate and Confers Glyphosate Resistance in Echinochloa colona[J]. Plant Physiol, 2019, 181(4): 1519-1534.
- ↑ http://2016.igem.org/Team:TJUSLS_China
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 294
Illegal BamHI site found at 2210 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 513
Illegal AgeI site found at 583
Illegal AgeI site found at 660 - 1000COMPATIBLE WITH RFC[1000]
