Difference between revisions of "Part:BBa K3338002"
Jonas Scholz (Talk | contribs) (→Part Design) |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
<partinfo>BBa_K3338002 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3338002 SequenceAndFeatures</partinfo> | ||
| − | |||
| − | |||
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 19: | Line 16: | ||
<!-- --> | <!-- --> | ||
| − | + | =Characterization= | |
| − | = | + | |
| − | + | ||
| − | + | ||
===Motivation=== | ===Motivation=== | ||
One of the central aspects of our inflammatory toxin sensor is the LPS-sensitivity of its promoter. Because of that, one of our goals was to design a LPS-sensitive promoter showing high induction following LPS-treatment. In a previous study it was shown that the human CMV immediate‐early (CMV-IE) gene enhancer/promoter shows NF-κB and c-Jun dependent regulation in response to LPS and bacterial CpG‐oligodeoxynucleotides (Lee <i>et al.</i> 2004). The experiments were performed in the macrophage cell line RAW 264.7 and showed that a maximum of CMV induction was reached 6\,h after LPS-supplementation followed by a decrease in promoter activity (Lee <i>et al.</i> 2004). This observation was not reproducible in HeLa-cells using a CMV-hGLuc construct (<html><a href="https://parts.igem.org/Part:BBa_K3338017">BBa_K3338017</a></html>) in the mammalian expression vector pEGFP-C2 (see figure 1). | One of the central aspects of our inflammatory toxin sensor is the LPS-sensitivity of its promoter. Because of that, one of our goals was to design a LPS-sensitive promoter showing high induction following LPS-treatment. In a previous study it was shown that the human CMV immediate‐early (CMV-IE) gene enhancer/promoter shows NF-κB and c-Jun dependent regulation in response to LPS and bacterial CpG‐oligodeoxynucleotides (Lee <i>et al.</i> 2004). The experiments were performed in the macrophage cell line RAW 264.7 and showed that a maximum of CMV induction was reached 6\,h after LPS-supplementation followed by a decrease in promoter activity (Lee <i>et al.</i> 2004). This observation was not reproducible in HeLa-cells using a CMV-hGLuc construct (<html><a href="https://parts.igem.org/Part:BBa_K3338017">BBa_K3338017</a></html>) in the mammalian expression vector pEGFP-C2 (see figure 1). | ||
| − | T--Hannover--results_promoter_CMV | + | |
| + | |||
| + | |||
| + | <html> | ||
| + | <img src="https://2020.igem.org/wiki/images/5/53/T--Hannover--results_promoter_CMV.png" class="center"> | ||
| + | </html> | ||
| + | Figure 1: Relative activity of CMV promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk. | ||
| + | |||
| + | |||
| + | |||
To achieve high induction of the CMV-promoter following LPS-treatment of the cells, we had to modify the promoter/enhancer-regions of the CMV-promoter. | To achieve high induction of the CMV-promoter following LPS-treatment of the cells, we had to modify the promoter/enhancer-regions of the CMV-promoter. | ||
===Part Design=== | ===Part Design=== | ||
| − | LPS-sensitivity of the CMV-promoter is mediated through NF-κB and AP1 transcription factor binding sites (Lee <i>et al.</i> 2004). In order to abolish effects of other transcription factors on promoter activity, the CMV-enhancer and large parts of the promoter were removed just leaving a minimal CMV promoter with the TATA-box and the initiator-sequence which are essential for promoter activity. To make the promoter sensing, we added AP1 and NF-κB transcription factor binding sites right in front of the CMV<sub>min</sub> promoter. These sequences were generated using a MATLAB tool which creates random AP1 and NF-κB sites based on the consensus sequence “GGGRNYYYCC” for NF-κB and “TGAXTCA” for AP1, where Y is C or T, R is A or G, X is C or G and N is any base (Chen <i>et al.</i> 1998, Alam and Den 1992). | + | LPS-sensitivity of the CMV-promoter is mediated through NF-κB and AP1 transcription factor binding sites (Lee <i>et al.</i> 2004). In order to abolish effects of other transcription factors on promoter activity, the CMV-enhancer and large parts of the promoter were removed just leaving a minimal CMV promoter with the TATA-box and the initiator-sequence which are essential for promoter activity. To make the promoter sensing, we added AP1 and NF-κB transcription factor binding sites right in front of the CMV<sub>min</sub> promoter. These sequences were generated using a MATLAB tool which creates random AP1 and NF-κB sites based on the consensus sequence “GGGRNYYYCC” for NF-κB and “TGAXTCA” for AP1, where Y is C or T, R is A or G, X is C or G and N is any base (Chen <i>et al.</i> 1998, Alam and Den 1992). The sequence consists of three NF-κB/AP-1-sites (see figure 2). |
| − | T--Hannover-- | + | |
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/5/56/T--Hannover--parts_synthp2.png" class="center" style="width: 95%; height: 95%"> | ||
| + | </p> | ||
| + | </html> | ||
| + | Figure 2: Schematic representation of the generation of the NF-κB-AP1-minCMV promoter_2. Combination of both transcription factor binding site sequences (3 times NF-κB/AP-1) and the minimal CMV promoter (A) yielded the synthetic promoter sequence (B). | ||
===Results=== | ===Results=== | ||
| − | In order to test the synthetic | + | In order to test the synthetic promoter described above, its activity following LPS-stimulation was ascertained using hGLuc as a reporter gene. Therefore SynthP_2 was cloned into pEGFP-C2 (<html><a href=" https://parts.igem.org/Part:BBa_K3338020">BBa_K3338020</a></html>) using AseI/HindIII to linearize the vector (CMV-enhancer/promoter and EGFP were removed). The constructs were transfected in HeLa cells. The cells were treated with LPS for 3 h and hGLuc expression was assessed 24 h and 48 h after LPS-treatment. Figure 3 shows quantitative data of the experiments. |
| − | T--Hannover-- | + | |
| + | |||
| + | |||
| + | <html> | ||
| + | <img src="https://2020.igem.org/wiki/images/f/fe/T--Hannover--results_promoter_minCMV_2.png" class="center"> | ||
| + | </html> | ||
| + | Figure 3: Relative activity of NF-κB-AP1-minCMV2 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk. | ||
| + | |||
| + | |||
| + | |||
| + | For the synthetic promoters a significant upregulation of the promoter activity was observed 24 h and 48 h after LPS-treatment of the cells for 3 h. The activity rises up to approximately 100 % over the basal activity. The highest values were observed for cells treated with 2 μg/mL LPS. Furthermore, the basal activities of different promoters was compared using the basal activity of the CMV promoter as a standard (see figure 4). | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <img src="https://2020.igem.org/wiki/images/9/93/T--Hannover--results_promoter_basal.png" class="center"> | ||
| + | </html> | ||
| + | Figure 4: Relative basal activity of all tested promoters in HeLa cells 48 hours (A) and 72 hours (B) after transfection. Data was normalized to CMV promoter which served as reference. Data shown represents mean ± SEM of n=4 biological replicates. | ||
| + | |||
| − | |||
| − | + | The results indicate that the synthetic promoter exhibits a higher basal activity than the original CMV-promoter. This could be due to higher transfection efficiency as compared to the original CMV-hGLuc construct because of the lesser plasmid size. However, further experiments are needed to clarify. The naturally occurring LPS-sensitive human IL-6 promoter shows a basal activity comparable with the CMV-promoter. For the usage as a LPS-sensor the promoter ideally exhibits a low basal activity like the IL-6 promoter. But even more important is its LPS sensitivity. Figure 5 indicates that the gain of IL-6 activity is significant but much smaller compared to the synthetic promoter. | |
| − | |||
| − | |||
| − | T--Hannover--results_promoter_IL6 | + | <html> |
| + | <img src="https://2020.igem.org/wiki/images/d/de/T--Hannover--results_promoter_IL6.png" class="center"> | ||
| + | </html> | ||
| + | Figure 5: Relative activity of IL6 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk. | ||
===Conclusions=== | ===Conclusions=== | ||
| − | Here we describe | + | Here we describe “synthetic promoter_2 with NF-κB and AP1 binding sites” as an improved CMV-promoter that is capable of detecting LPS in the surrounding of the cell via the TLR4-NF-κB/c-Jun pathway leading to enhanced gene expression. It is much more sensitive than the naturally occurring LPS-sensitive interleukin-6 promoter but shows a higher basal activity than the IL-6 and the CMV-promoter. Because of that, it can on the one hand used as an CMV-based LPS-sensor but on the other hand also as an improved version of the natural CMV-promoter regarding promoter strength. |
=References= | =References= | ||
| Line 57: | Line 84: | ||
Chen, F. E., Huang, D. B., Chen, Y. Q., & Ghosh, G. (1998). Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. <i>Nature</i>, 391(6665), 410–413. | Chen, F. E., Huang, D. B., Chen, Y. Q., & Ghosh, G. (1998). Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. <i>Nature</i>, 391(6665), 410–413. | ||
| − | Lee, Y., Sohn, W. J., Kim, D. S., & Kwon, H. J. (2004). NF-kappaB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. | + | Lee, Y., Sohn, W. J., Kim, D. S., & Kwon, H. J. (2004). NF-kappaB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. <i>European journal of biochemistry</i>, 271(6), 1094–1105. |
Latest revision as of 16:53, 27 October 2020
Synthetic promoter_2 with NF-κB and AP1 binding sites
Usage and Biology
The presented synthetic promoter is able to recognize LPS in the surrounding of the cell via the Toll-like receptor, NF-κB/AP1 pathway leading to strong induction of gene expression. Due to its relatively low basal activity and strong induction following LPS supplementation it is perfectly suitable for the application as a LPS-sensor. It was designed as an improvement of the CMV promoter making it LPS-sensing. This improvment is also documented on the wiki page of the original part BBa_I712004. The novel synthetic promoter is based on a minimal CMV-promoter just containing the TATA-box and the Initiator-Sequence of the original CMV-promoter. Upstream of the minimal Promoter three repetitions of randomly generated AP1-NF-κB binding sites are localized.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1
Illegal NotI site found at 58 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Motivation
One of the central aspects of our inflammatory toxin sensor is the LPS-sensitivity of its promoter. Because of that, one of our goals was to design a LPS-sensitive promoter showing high induction following LPS-treatment. In a previous study it was shown that the human CMV immediate‐early (CMV-IE) gene enhancer/promoter shows NF-κB and c-Jun dependent regulation in response to LPS and bacterial CpG‐oligodeoxynucleotides (Lee et al. 2004). The experiments were performed in the macrophage cell line RAW 264.7 and showed that a maximum of CMV induction was reached 6\,h after LPS-supplementation followed by a decrease in promoter activity (Lee et al. 2004). This observation was not reproducible in HeLa-cells using a CMV-hGLuc construct (BBa_K3338017) in the mammalian expression vector pEGFP-C2 (see figure 1).
 Figure 1: Relative activity of CMV promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
Figure 1: Relative activity of CMV promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
To achieve high induction of the CMV-promoter following LPS-treatment of the cells, we had to modify the promoter/enhancer-regions of the CMV-promoter.
Part Design
LPS-sensitivity of the CMV-promoter is mediated through NF-κB and AP1 transcription factor binding sites (Lee et al. 2004). In order to abolish effects of other transcription factors on promoter activity, the CMV-enhancer and large parts of the promoter were removed just leaving a minimal CMV promoter with the TATA-box and the initiator-sequence which are essential for promoter activity. To make the promoter sensing, we added AP1 and NF-κB transcription factor binding sites right in front of the CMVmin promoter. These sequences were generated using a MATLAB tool which creates random AP1 and NF-κB sites based on the consensus sequence “GGGRNYYYCC” for NF-κB and “TGAXTCA” for AP1, where Y is C or T, R is A or G, X is C or G and N is any base (Chen et al. 1998, Alam and Den 1992). The sequence consists of three NF-κB/AP-1-sites (see figure 2).

Results
In order to test the synthetic promoter described above, its activity following LPS-stimulation was ascertained using hGLuc as a reporter gene. Therefore SynthP_2 was cloned into pEGFP-C2 (BBa_K3338020) using AseI/HindIII to linearize the vector (CMV-enhancer/promoter and EGFP were removed). The constructs were transfected in HeLa cells. The cells were treated with LPS for 3 h and hGLuc expression was assessed 24 h and 48 h after LPS-treatment. Figure 3 shows quantitative data of the experiments.
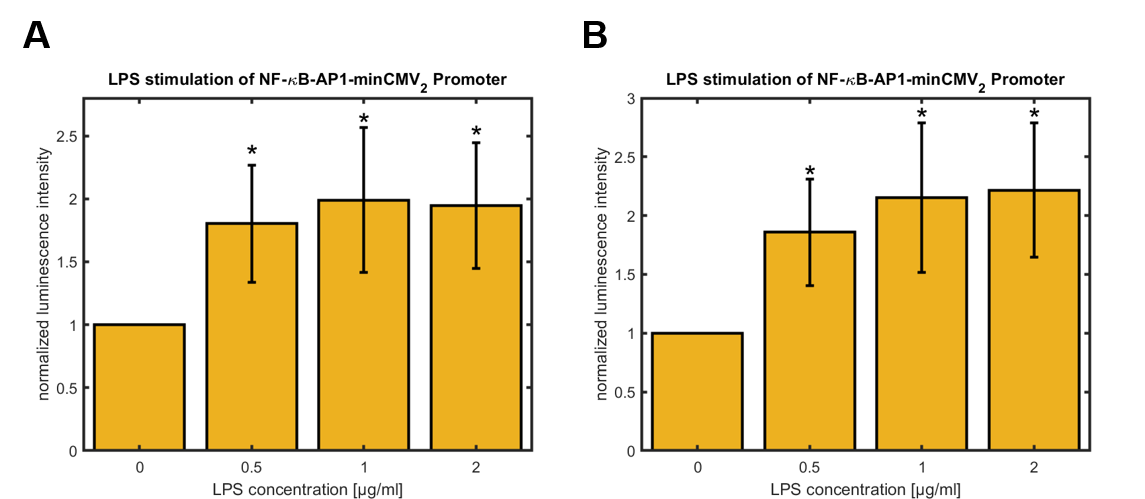 Figure 3: Relative activity of NF-κB-AP1-minCMV2 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
Figure 3: Relative activity of NF-κB-AP1-minCMV2 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
For the synthetic promoters a significant upregulation of the promoter activity was observed 24 h and 48 h after LPS-treatment of the cells for 3 h. The activity rises up to approximately 100 % over the basal activity. The highest values were observed for cells treated with 2 μg/mL LPS. Furthermore, the basal activities of different promoters was compared using the basal activity of the CMV promoter as a standard (see figure 4).
 Figure 4: Relative basal activity of all tested promoters in HeLa cells 48 hours (A) and 72 hours (B) after transfection. Data was normalized to CMV promoter which served as reference. Data shown represents mean ± SEM of n=4 biological replicates.
Figure 4: Relative basal activity of all tested promoters in HeLa cells 48 hours (A) and 72 hours (B) after transfection. Data was normalized to CMV promoter which served as reference. Data shown represents mean ± SEM of n=4 biological replicates.
The results indicate that the synthetic promoter exhibits a higher basal activity than the original CMV-promoter. This could be due to higher transfection efficiency as compared to the original CMV-hGLuc construct because of the lesser plasmid size. However, further experiments are needed to clarify. The naturally occurring LPS-sensitive human IL-6 promoter shows a basal activity comparable with the CMV-promoter. For the usage as a LPS-sensor the promoter ideally exhibits a low basal activity like the IL-6 promoter. But even more important is its LPS sensitivity. Figure 5 indicates that the gain of IL-6 activity is significant but much smaller compared to the synthetic promoter.
 Figure 5: Relative activity of IL6 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
Figure 5: Relative activity of IL6 promoter in HeLa cells 24 hours (A) and 48 hours (B) after treatment with different concentrations of LPS for 3 hours, normalized to untreated control (0 µg/mL LPS). Data shown represents mean ± SEM of n=4 biological replicates. Statistical analysis was performed by unpaired t-test in comparison to untreated control, significance level: 10 %, significance is indicated by asterisk.
Conclusions
Here we describe “synthetic promoter_2 with NF-κB and AP1 binding sites” as an improved CMV-promoter that is capable of detecting LPS in the surrounding of the cell via the TLR4-NF-κB/c-Jun pathway leading to enhanced gene expression. It is much more sensitive than the naturally occurring LPS-sensitive interleukin-6 promoter but shows a higher basal activity than the IL-6 and the CMV-promoter. Because of that, it can on the one hand used as an CMV-based LPS-sensor but on the other hand also as an improved version of the natural CMV-promoter regarding promoter strength.
References
Alam, J., & Den, Z. (1992). Distal AP-1 binding sites mediate basal level enhancement and TPA induction of the mouse heme oxygenase-1 gene. The Journal of biological chemistry, 267(30), 21894–21900.
Chen, F. E., Huang, D. B., Chen, Y. Q., & Ghosh, G. (1998). Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature, 391(6665), 410–413.
Lee, Y., Sohn, W. J., Kim, D. S., & Kwon, H. J. (2004). NF-kappaB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. European journal of biochemistry, 271(6), 1094–1105.
