Difference between revisions of "Part:BBa K3429011"
| (8 intermediate revisions by the same user not shown) | |||
| Line 175: | Line 175: | ||
<a class="referencestd" href="https://doi.org/10.1038/nprot.2013.074" target="_blank">[1] Steven A. Combs <i>et al.</i>, Small-molecule ligand docking into comparative models with Rosetta, Nature Protocols, 2013, 8:1277–1298, doi:10.1038/nprot.2013.074</a><br></div> | <a class="referencestd" href="https://doi.org/10.1038/nprot.2013.074" target="_blank">[1] Steven A. Combs <i>et al.</i>, Small-molecule ligand docking into comparative models with Rosetta, Nature Protocols, 2013, 8:1277–1298, doi:10.1038/nprot.2013.074</a><br></div> | ||
</html> | </html> | ||
| + | |||
| + | |||
| + | |||
| + | ==Contribution: Team Aboa 2021== | ||
| + | <!-- | ||
| + | Group: Team Aboa 2021 | ||
| + | Author: Juuli Hietarinne | ||
| + | Summary: We added some results we obtained using this Part. | ||
| + | <!-- --> | ||
| + | |||
| + | |||
| + | |||
| + | ===Protein production and purification=== | ||
| + | |||
| + | [[Image:T--Aboa--result1.jpg|thumb|400px|<b>Figure 4. The verification digestions of the pET36b-cotA, pET36b-cueO and pET36b-yak expression constructs.</b> The expression constructs were digested with NdeI and XhoI restriction enzymes. The 1 kb DNA ladder (NEB) was used as a molecular weight standard, the DNA was colored with SyBR Safe DNA Gel Stain (ThermoFisher), and the gel was visualized under UV light. The agarose gel used was 1.2 %. | ||
| + | ]] | ||
| + | |||
| + | We transformed <i>E. coli</i> BL21(DE3) cells with pET36b-<i>cotA</i> expression constructs. Three colonies from the strain were analyzed by restriction digestion with NdeI and XhoI. They were true transformants, as seen in the Figure 4. The protein production of the cells was induced by IPTG, the cells were lysed by sonication and His-tagged laccases were extracted with nickel affinity chromatography. The sizes of purified proteins were verified with SDS-PAGE, which showed clear bands around 65 kDa, expectedly corresponding to the calculated size of CotA (Fig. 5). That suggests that CotA was successfully produced by generated <i>E. coli</i> BL21(DE3) strains. | ||
| + | |||
| + | |||
| + | [[Image:T--Aboa--fig3.jpg|thumb|400px|<b>Figure 5. SDS-PAGE of raw extracts and purified enzyme fractions.</b> Bio-Rad Ready Gel® Precast Polyacrylamide Gel was used and the ladder was the PageRuler Unstained Protein Ladder (Thermo Fisher). | ||
| + | ]] | ||
| + | |||
| + | |||
| + | |||
| + | ===Characterization and Results=== | ||
| + | |||
| + | To measure the laccase activity, we performed a kinetic laccase assay using syringaldazine as substrate. The reaction is based on substrate oxidation which can be spectrophotometrically monitored as signal increase at 530 nm. It was clearly seen that CotA catalyzed substrate oxidation as seen in increasing progress curves (Fig. 6). While the signal of the control remained near the baseline, other progress curve slopes seemed to correlate with the CotA concentrations. The 20 mM reaction profiles using 5 µl of the CotA fraction were identical between graphs, showing that the result was reproducible (Fig. 6A-B). | ||
| + | |||
| + | [[Image:T--Aboa--fig5.jpg|thumb|900px|<b>Figure 6. Representative kinetic activity assays with the purified CotA laccase fractions and syringaldazine measured at 530 nm.</b> A) Reactions with 20 mM syringaldazine and 0 µl (green line), 1 µl (red line), 5 µl (blue line) and 10 µl (purple line) CotA purification fraction. B) Reactions with 5 µl CotA and varying concentrations of the syringaldazine: 0 mM (green line), 5 mM (red line), 20 mM (blue line).]] | ||
| + | |||
| + | |||
| + | This confirmed that the laccase CotA was catalytically active and capable of oxidizing syringaldazine. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | |||
Latest revision as of 15:09, 21 October 2021
Laccase CotA
Profile
| Name | CotA-Laccase (Spore coat protein A) |
| Base pairs | 1542 |
| Molecular weight | 58.51 kDa |
| Origin | Bacillus subtilis, synthetic |
| Properties | CotA laccase from B. subtilis is a copper dependant oxireductase with a broad substrate variety of polyphenolic substrates such as diclofenac and ABTS and possess a T1 copper for primary electron transfer to the substrate and a T2/3 Cu-cluster. |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
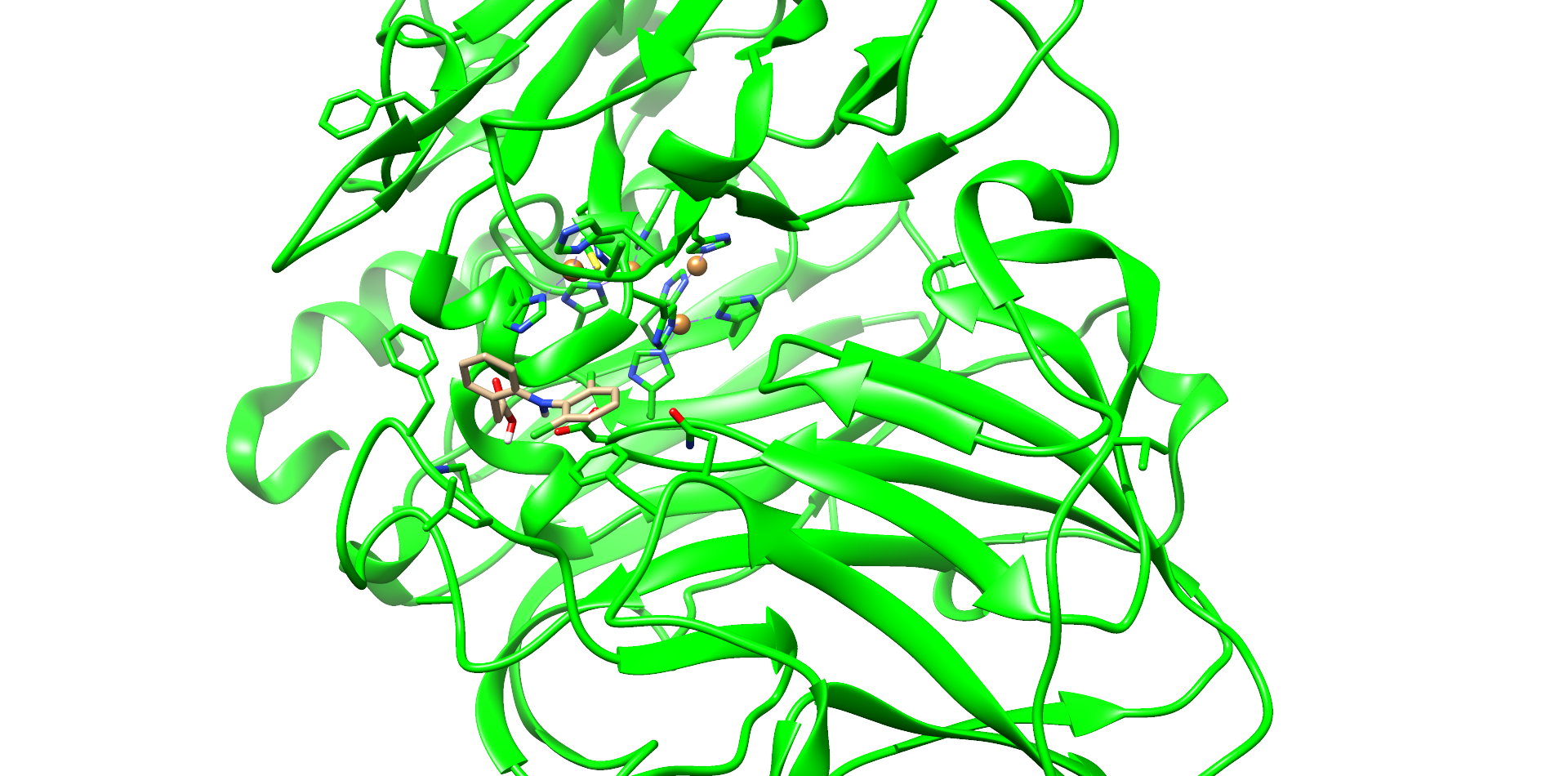
CotA Docking
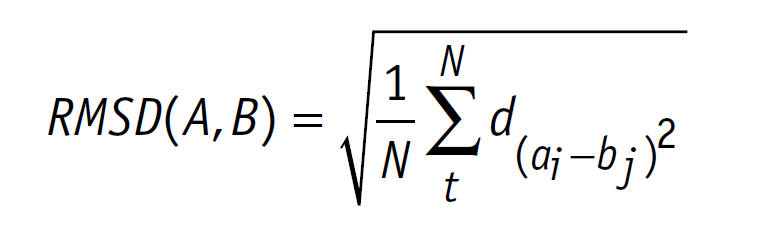

| Enzyme |
Substance |
Best Interface score |
Best Total score |
|---|---|---|---|
| CotA |
4-Nonylphenol |
-11.014 |
-1232.434 |
| Ibuprofen |
-12.170 |
-1232.533 |
|
| Carbamazepin |
-12.633 |
-1230.243 |
|
| Diclofenac |
-12.820 |
-1230.488 |
|
| T. Versicolor |
Diclofenac |
-13.266 |
-1528.821 |
Contribution: Team Aboa 2021
Protein production and purification

We transformed E. coli BL21(DE3) cells with pET36b-cotA expression constructs. Three colonies from the strain were analyzed by restriction digestion with NdeI and XhoI. They were true transformants, as seen in the Figure 4. The protein production of the cells was induced by IPTG, the cells were lysed by sonication and His-tagged laccases were extracted with nickel affinity chromatography. The sizes of purified proteins were verified with SDS-PAGE, which showed clear bands around 65 kDa, expectedly corresponding to the calculated size of CotA (Fig. 5). That suggests that CotA was successfully produced by generated E. coli BL21(DE3) strains.
Characterization and Results
To measure the laccase activity, we performed a kinetic laccase assay using syringaldazine as substrate. The reaction is based on substrate oxidation which can be spectrophotometrically monitored as signal increase at 530 nm. It was clearly seen that CotA catalyzed substrate oxidation as seen in increasing progress curves (Fig. 6). While the signal of the control remained near the baseline, other progress curve slopes seemed to correlate with the CotA concentrations. The 20 mM reaction profiles using 5 µl of the CotA fraction were identical between graphs, showing that the result was reproducible (Fig. 6A-B).

This confirmed that the laccase CotA was catalytically active and capable of oxidizing syringaldazine.