Difference between revisions of "Part:BBa K3617007"
| (12 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
==<partinfo>BBa_K3617007 short</partinfo>== | ==<partinfo>BBa_K3617007 short</partinfo>== | ||
| − | + | This biobrick is a membrane bound transcription factor designed to activate upon cleavage by a TEV protease. Similar to BBa_K165020 it is composed of the lexA DNA binding domain from <i> Escherichia coli </i> and an activation domain from the herpes simplex virus VP16. As an improvement of BBa_K165020, this biobrick contains a transmembrane domain linked by a flexible link containing a TEV protease cleavage site. The transcription factor is by default inactive, however upon cleavage by the TEV protease the transcription factor is released and transported to the nucleus where it activates transcription. | |
==Usage== | ==Usage== | ||
| − | The biobrick | + | The biobrick was designed to function as an accessory protein in a series of <i> S. cerevisiae </i> interleukin biosensors. It was expressed together with two modular receptor proteins also located at the plasma membrane. The receptors had human interleukin receptors extracellularly and either the C- or N-terminal part of a split TEV protease intracellularly. Split TEV protease is a TEV protease divided into two halves, which only regains its proteolytic activity when the halves are complemented. The idea was that when the extracellular parts of the interleukin receptor proteins bound to the target interleukin, the receptors would come together, causing the split TEV protease to be complemented and regain proteolytic activity. One split TEV protease could then cleave multiple copies of the biobrick leading to amplification and transduction of the signal to the nucleus. |
==<span class='h3bb'>Sequence and Features</span>== | ==<span class='h3bb'>Sequence and Features</span>== | ||
<partinfo>BBa_K3617007 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3617007 SequenceAndFeatures</partinfo> | ||
| − | This biobrick consists of multiple parts; An | + | This biobrick consists of multiple parts; An endoplasmic reticulum import signal peptide from the <i> Saccharomyces cerevisiae </i> cell wall integrity and stress response component 1 (Wsc1). The transmembrane domain of Wsc1, as well as the transcriptional activation domain of VP16 from herpes simplex virus linked to the LexA DNA binding domain from <i> Escherichia coli</i>. The transmembrane domain and synthetic transcription factor are linked by a flexible GS linker followed by a seven amino acid consensus sequence (ENLYFQ’G) for the cut site for Tobacco Etch Virus Nuclear Inclusion protein a (TEV protease). |
==Sequence optimization== | ==Sequence optimization== | ||
| − | The sequence was codon optimized for S. cerevisiae. The recognition sequences for SpeI, XbaI, NotI, EcoRI, PstI were avoided to follow the RFC10 standard. | + | The sequence was codon optimized for <i> S. cerevisiae</i>. The recognition sequences for SpeI, XbaI, NotI, EcoRI, PstI were avoided to follow the RFC10 standard. |
| − | + | ||
==Structure and function== | ==Structure and function== | ||
| − | [[Image:T--UCopenhagen--TMDesign.png| | + | [[Image:T--UCopenhagen--TMDesign.png|450px|thumb|left|<p align="justify"> '''Figure 1: Mechanism for LexA-VP16-mediated gene expression via split-TEV protease cleavage.''' Split-TEV protease complementation leading to cleavage of a TEV-recognition sequence near LexA-VP16, thereby releasing LexA-VP16 to initiate gene expression. As the TEV protease is able to cleave more than one membrane-bound transcription factor, the signal is further amplified. </p>]] |
| − | + | Both the signal peptide and transmembrane domain stem from the S. cerevisiae cell wall integrity sensor Wsc1. As described by Ivanusic et al. 2015, they are commonly used in yeast-two-hybrid assays of membrane protein-protein interactions as a way to increase the likelihood of plasma membrane localization of proteins. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Between the transmembrane domain and the LexA-VP16 transcription factor is a flexible linker with a TEV protease cleavage site. Upon cleavage the transcription factor to localize to the nucleus, where it activates the transcription of genes downstream from its associated promoter. | ||
| + | The LexA DNA binding domain of the synthetic transcription factor binds to the lexAop promoter which is incorporated into a synthetic promoter consisting of six lexAop sequences and the core promoter from the eno1 promoter endogenous to <i> S. cerevisiae</i>. Since the transcription factor does not regulate endogenous gene expression in <i> S. cerevisiae</i>, possible crosstalk is minimized. | ||
==Cleavage by inducible TEV protease== | ==Cleavage by inducible TEV protease== | ||
| − | In order to test | + | In order to test the functionality of the biobrick it was expressed in S. cerevisiae together with a TEV protease under the control of the galactose inducible scGAL1 promoter, and the NanoLuc luciferase reporter, an enhanced version of luciferase, under control of the (6x)lexAop-eno1 promoter. In the absence of galactose, no TEV protease should be present, and the LexA-VP16 transcription factor should remain membrane-bound. However, in the presence of galactose expression of the TEV protease would be initiated resulting in cleavage of the linker region and release of the LexA-VP16 transcription factor. |
| − | The strain was | + | The strain was incubated in both the presence and absence of galactose. Subsequently, proteins were extracted from the cell cultures using YeastBuster, an industrial protein extraction reagent for a fast extract of active proteins from yeast without mechanical disruption or enzymatic lysis, and a luminescence assay was performed in order to analyze luciferase expression (Figure 2). |
| − | [[Image:T--UCopenhagen--results-TMD-TFassay.jpg|700px|thumb|center|<p align="justify"> ''' | + | [[Image:T--UCopenhagen--results-TMD-TFassay.jpg|700px|thumb|center|<p align="justify"> '''Figure 2: Luminescence assay based on cleavage of membrane-bound transcription factor.''' Cells were expressed without (glc) and with (gal+raf) induction of the TEV protease. A 700-fold increase in luminescence was seen upon induction of the TEV protease. Data is the result of three biological replicates.</p>]] |
| − | + | As seen in the figure, a 700-fold increase in luminescence was seen after induction of the TEV protease. This strongly suggests that the biobrick works as intended. However, we cannot exclude that part of the difference in luciferase concentrations stems from other effects of changing the media. | |
A 700-fold increase in luminescence intensity was observed following expression of the TEV protease. This result infers that the biobrick is working as intended. | A 700-fold increase in luminescence intensity was observed following expression of the TEV protease. This result infers that the biobrick is working as intended. | ||
| Line 48: | Line 43: | ||
<partinfo>BBa_K3617007 parameters</partinfo> | <partinfo>BBa_K3617007 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ==References== | ||
| + | <p>[1] Ivanusic, Daniel, Jürgen J. Heinisch, Magdalena Eschricht, Ulrike Laube, and Joachim Denner. 2015. “Improved Split-Ubiquitin Screening Technique to Identify Surface Membrane Protein-Protein Interactions.” BioTechniques. https://doi.org/10.2144/000114315. | ||
Latest revision as of 02:05, 28 October 2020
TMDWsc1-TEV_recseq-LexA-VP16
This biobrick is a membrane bound transcription factor designed to activate upon cleavage by a TEV protease. Similar to BBa_K165020 it is composed of the lexA DNA binding domain from Escherichia coli and an activation domain from the herpes simplex virus VP16. As an improvement of BBa_K165020, this biobrick contains a transmembrane domain linked by a flexible link containing a TEV protease cleavage site. The transcription factor is by default inactive, however upon cleavage by the TEV protease the transcription factor is released and transported to the nucleus where it activates transcription.
Usage
The biobrick was designed to function as an accessory protein in a series of S. cerevisiae interleukin biosensors. It was expressed together with two modular receptor proteins also located at the plasma membrane. The receptors had human interleukin receptors extracellularly and either the C- or N-terminal part of a split TEV protease intracellularly. Split TEV protease is a TEV protease divided into two halves, which only regains its proteolytic activity when the halves are complemented. The idea was that when the extracellular parts of the interleukin receptor proteins bound to the target interleukin, the receptors would come together, causing the split TEV protease to be complemented and regain proteolytic activity. One split TEV protease could then cleave multiple copies of the biobrick leading to amplification and transduction of the signal to the nucleus.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 507
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
This biobrick consists of multiple parts; An endoplasmic reticulum import signal peptide from the Saccharomyces cerevisiae cell wall integrity and stress response component 1 (Wsc1). The transmembrane domain of Wsc1, as well as the transcriptional activation domain of VP16 from herpes simplex virus linked to the LexA DNA binding domain from Escherichia coli. The transmembrane domain and synthetic transcription factor are linked by a flexible GS linker followed by a seven amino acid consensus sequence (ENLYFQ’G) for the cut site for Tobacco Etch Virus Nuclear Inclusion protein a (TEV protease).
Sequence optimization
The sequence was codon optimized for S. cerevisiae. The recognition sequences for SpeI, XbaI, NotI, EcoRI, PstI were avoided to follow the RFC10 standard.
Structure and function

Figure 1: Mechanism for LexA-VP16-mediated gene expression via split-TEV protease cleavage. Split-TEV protease complementation leading to cleavage of a TEV-recognition sequence near LexA-VP16, thereby releasing LexA-VP16 to initiate gene expression. As the TEV protease is able to cleave more than one membrane-bound transcription factor, the signal is further amplified.
Both the signal peptide and transmembrane domain stem from the S. cerevisiae cell wall integrity sensor Wsc1. As described by Ivanusic et al. 2015, they are commonly used in yeast-two-hybrid assays of membrane protein-protein interactions as a way to increase the likelihood of plasma membrane localization of proteins.
Between the transmembrane domain and the LexA-VP16 transcription factor is a flexible linker with a TEV protease cleavage site. Upon cleavage the transcription factor to localize to the nucleus, where it activates the transcription of genes downstream from its associated promoter.
The LexA DNA binding domain of the synthetic transcription factor binds to the lexAop promoter which is incorporated into a synthetic promoter consisting of six lexAop sequences and the core promoter from the eno1 promoter endogenous to S. cerevisiae. Since the transcription factor does not regulate endogenous gene expression in S. cerevisiae, possible crosstalk is minimized.
Cleavage by inducible TEV protease
In order to test the functionality of the biobrick it was expressed in S. cerevisiae together with a TEV protease under the control of the galactose inducible scGAL1 promoter, and the NanoLuc luciferase reporter, an enhanced version of luciferase, under control of the (6x)lexAop-eno1 promoter. In the absence of galactose, no TEV protease should be present, and the LexA-VP16 transcription factor should remain membrane-bound. However, in the presence of galactose expression of the TEV protease would be initiated resulting in cleavage of the linker region and release of the LexA-VP16 transcription factor.
The strain was incubated in both the presence and absence of galactose. Subsequently, proteins were extracted from the cell cultures using YeastBuster, an industrial protein extraction reagent for a fast extract of active proteins from yeast without mechanical disruption or enzymatic lysis, and a luminescence assay was performed in order to analyze luciferase expression (Figure 2).
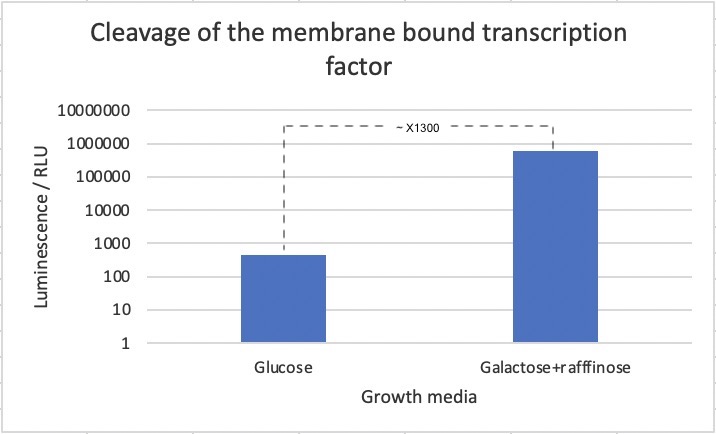
Figure 2: Luminescence assay based on cleavage of membrane-bound transcription factor. Cells were expressed without (glc) and with (gal+raf) induction of the TEV protease. A 700-fold increase in luminescence was seen upon induction of the TEV protease. Data is the result of three biological replicates.
As seen in the figure, a 700-fold increase in luminescence was seen after induction of the TEV protease. This strongly suggests that the biobrick works as intended. However, we cannot exclude that part of the difference in luciferase concentrations stems from other effects of changing the media.
A 700-fold increase in luminescence intensity was observed following expression of the TEV protease. This result infers that the biobrick is working as intended.
References
[1] Ivanusic, Daniel, Jürgen J. Heinisch, Magdalena Eschricht, Ulrike Laube, and Joachim Denner. 2015. “Improved Split-Ubiquitin Screening Technique to Identify Surface Membrane Protein-Protein Interactions.” BioTechniques. https://doi.org/10.2144/000114315.
