Difference between revisions of "Part:BBa K3352006"
| (6 intermediate revisions by 3 users not shown) | |||
| Line 7: | Line 7: | ||
<b><font size="+1.2"> Engineering Success </font></b> | <b><font size="+1.2"> Engineering Success </font></b> | ||
| − | We added the new part BBa_K3352000 into the iGEM registry. The SplintR ligase coding sequence was obtained from the amino acid sequence in Paramecium bursaria Chlorella virus 1 (PBCV-1) [1]. Optimizing this sequence for E. coli expression and removing the PstI cutting site, we attached a 6x histidine tag (6x his-tag) upstream of the SplintR ligase sequence through a glycine-serine linker (GS linker). | + | We added the new part BBa_K3352000 into the iGEM registry. The SplintR ligase coding sequence was obtained from the amino acid sequence in Paramecium bursaria Chlorella virus 1 (PBCV-1) [1]. Optimizing this sequence for <i>E. coli</i> expression and removing the PstI cutting site, we attached a 6x histidine tag (6x his-tag) upstream of the SplintR ligase sequence through a glycine-serine linker (GS linker). |
https://2020.igem.org/wiki/images/thumb/a/a6/T--TAS_Taipei--Parts_2000.PNG/800px-T--TAS_Taipei--Parts_2000.PNG | https://2020.igem.org/wiki/images/thumb/a/a6/T--TAS_Taipei--Parts_2000.PNG/800px-T--TAS_Taipei--Parts_2000.PNG | ||
| Line 19: | Line 19: | ||
https://2020.igem.org/wiki/images/thumb/d/da/T--TAS_Taipei--Experimental_2.png/800px-T--TAS_Taipei--Experimental_2.png | https://2020.igem.org/wiki/images/thumb/d/da/T--TAS_Taipei--Experimental_2.png/800px-T--TAS_Taipei--Experimental_2.png | ||
| − | <b> Figure 2 | + | <b> Figure 2: Design of T7 Promoter SplintR ligase Expressing Construct (BBa_K3352006) </b> |
| − | Cloning this part into the iGEM ampicillin backbone pSB1A3, the plasmid was transformed into DH5⍺ E. coli cells for plasmid replication and subsequently miniprepped for a high yield of plasmid. We then transformed these plasmids into BL21(DE3) E. coli cells. Growing an overnight culture and measuring the OD600 to dilute cells to standardized populations, we induced expression with 0.1M IPTG once the OD600 surpassed 0.5. Liquid cultures were grown for an additional 2 hours. | + | Cloning this part into the iGEM ampicillin backbone pSB1A3, the plasmid was transformed into DH5⍺ <i>E. coli</i> cells for plasmid replication and subsequently miniprepped for a high yield of plasmid. We then transformed these plasmids into BL21(DE3) <i>E. coli</i> cells. Growing an overnight culture and measuring the OD600 to dilute cells to standardized populations, we induced expression with 0.1M IPTG once the OD600 surpassed 0.5. Liquid cultures were grown for an additional 2 hours. |
After protein expression, we harvested and lysed the cells with xTractor Lysis Buffer and centrifuged them for sample preparation [5]. This was followed by protein purification through Ni sepharose affinity chromatography, which could isolate our his-tagged SplintR ligase enzymes. | After protein expression, we harvested and lysed the cells with xTractor Lysis Buffer and centrifuged them for sample preparation [5]. This was followed by protein purification through Ni sepharose affinity chromatography, which could isolate our his-tagged SplintR ligase enzymes. | ||
| Line 27: | Line 27: | ||
https://2020.igem.org/wiki/images/6/61/T--TAS_Taipei--purified2006.png | https://2020.igem.org/wiki/images/6/61/T--TAS_Taipei--purified2006.png | ||
| − | <b> Figure | + | <b> Figure 3: SDS-PAGE on purified proteins with the T7 promoter SplintR ligase expressing construct (BBa_K3352006). </b> |
Our results in Figure #3 shows a protein band at around 35.2kDa, which is the molecular weight of our SplintR ligase enzyme (containing the 6x His-tag and GS linker attached to the enzyme). This provides evidence that SplintR ligase was expressed and purified. | Our results in Figure #3 shows a protein band at around 35.2kDa, which is the molecular weight of our SplintR ligase enzyme (containing the 6x His-tag and GS linker attached to the enzyme). This provides evidence that SplintR ligase was expressed and purified. | ||
| Line 35: | Line 35: | ||
However, because SplintR ligase is stored in unfavorable elution buffer conditions with compounds such as imidazole, we conducted a dialysis buffer exchange. Using Vivaspin® 6 Centrifugal Concentrators, we further enriched our SplintR ligase sample [4]. The new buffer conditions contain 10mM Tris-Hcl, 300mM NaCl, 1mM DTT, 0.1mM EDTA, and 50% glycerol at a pH of 7.4, which is the optimal storage conditions and pH starting point for our RCA test. | However, because SplintR ligase is stored in unfavorable elution buffer conditions with compounds such as imidazole, we conducted a dialysis buffer exchange. Using Vivaspin® 6 Centrifugal Concentrators, we further enriched our SplintR ligase sample [4]. The new buffer conditions contain 10mM Tris-Hcl, 300mM NaCl, 1mM DTT, 0.1mM EDTA, and 50% glycerol at a pH of 7.4, which is the optimal storage conditions and pH starting point for our RCA test. | ||
| + | <html><body><video width="60%" controls><source src="https://2020.igem.org/wiki/images/4/45/T--TAS_Taipei--newmovie.mp4" type="video/mp4"></video></body></html> | ||
| − | + | <b> Video 1: SARS-CoV-2 RNA RCA Test with Purified SplintR ligase. </b> | |
| − | + | ||
| − | <b> Video | + | |
Conducting the RCA test on SARS-CoV-2, the PCR tube under letter b is a negative control with no SARS-CoV-2 RNA Target and the PCR tube under letter c is our test group with a SARS-CoV-2 RNA Target. Both tubes use our purified SplintR ligase. The test group yielded a color change from purple to orange in just one hour and ultimately ended at yellow while our negative control remained purple. This results indicates that our SplintR ligase enzymes enabled an accurate detection of the presence of SARS-CoV-2 through its synthetic RNA fragment in our RCA diagnostic test. | Conducting the RCA test on SARS-CoV-2, the PCR tube under letter b is a negative control with no SARS-CoV-2 RNA Target and the PCR tube under letter c is our test group with a SARS-CoV-2 RNA Target. Both tubes use our purified SplintR ligase. The test group yielded a color change from purple to orange in just one hour and ultimately ended at yellow while our negative control remained purple. This results indicates that our SplintR ligase enzymes enabled an accurate detection of the presence of SARS-CoV-2 through its synthetic RNA fragment in our RCA diagnostic test. | ||
| + | |||
https://2020.igem.org/wiki/images/thumb/b/bc/T--TAS_Taipei--Engineering_4.png/800px-T--TAS_Taipei--Engineering_4.png | https://2020.igem.org/wiki/images/thumb/b/bc/T--TAS_Taipei--Engineering_4.png/800px-T--TAS_Taipei--Engineering_4.png | ||
| − | <b> Figure | + | |
| + | <b> Figure 4: pH over Time Analysis of both tubes in the SARS-CoV-2 RNA RCA Test with Purified SplintR ligase </b> | ||
Using our modeling team’s software to transform our qualitative viral detection readout to quantitative data, we constructed a pH over time graph through the change in hues of both tubes in the video. As shown in Figure #4, there is only minimal fluctuation around the same pH for our negative control and a rapid significant drop in pH of our test group. This is expected due to the production of hydrogen ions from our RCA test that ultimately results in the color change. This provides further validation that SplintR ligase enzyme was expressed, purified, and functioned its intended use. | Using our modeling team’s software to transform our qualitative viral detection readout to quantitative data, we constructed a pH over time graph through the change in hues of both tubes in the video. As shown in Figure #4, there is only minimal fluctuation around the same pH for our negative control and a rapid significant drop in pH of our test group. This is expected due to the production of hydrogen ions from our RCA test that ultimately results in the color change. This provides further validation that SplintR ligase enzyme was expressed, purified, and functioned its intended use. | ||
Latest revision as of 03:15, 26 October 2020
T7 + RBS SplintR Ligase Expressing Construct
The composite part utilizes a T7 promoter(BBa_J64997), a ribosome binding site, SplintR Ligase(BBa_K3352000), and a double terminator.
Engineering Success
We added the new part BBa_K3352000 into the iGEM registry. The SplintR ligase coding sequence was obtained from the amino acid sequence in Paramecium bursaria Chlorella virus 1 (PBCV-1) [1]. Optimizing this sequence for E. coli expression and removing the PstI cutting site, we attached a 6x histidine tag (6x his-tag) upstream of the SplintR ligase sequence through a glycine-serine linker (GS linker).
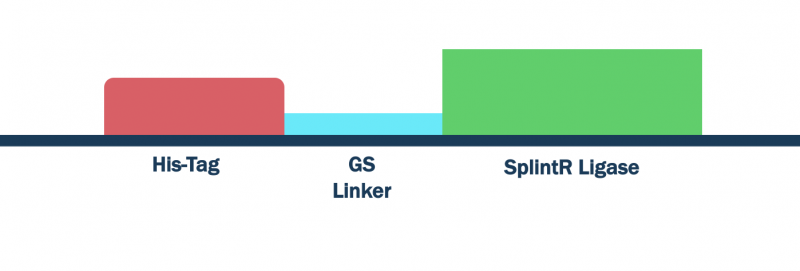
Figure 1: Open reading frame for SplintR ligase expression and isolation.
SplintR ligase catalyzes the ligation of adjacent single-stranded DNA splinted by complementary RNA strands [2]. Reported to proofread fully complementary in DNA/RNA hybrids down to the single nucleotide polymorph level, this enzyme could be an essential component to viral diagnostic testing [3]. In our project, it enables us to increase the specificity of our test by conducting direct RNA targeting.
In order to test this part, we flanked it with upstream T7 promoter (BBa_J64997), strong RBS (BBa_B0034) and downstream double terminator (BBa_B0015). This formed our composite part BBa_K3352006).
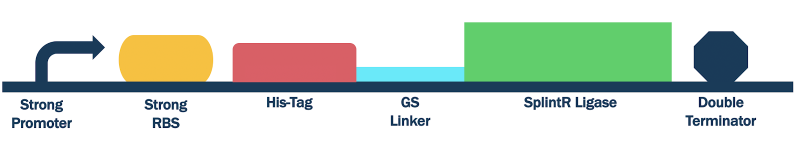
Figure 2: Design of T7 Promoter SplintR ligase Expressing Construct (BBa_K3352006)
Cloning this part into the iGEM ampicillin backbone pSB1A3, the plasmid was transformed into DH5⍺ E. coli cells for plasmid replication and subsequently miniprepped for a high yield of plasmid. We then transformed these plasmids into BL21(DE3) E. coli cells. Growing an overnight culture and measuring the OD600 to dilute cells to standardized populations, we induced expression with 0.1M IPTG once the OD600 surpassed 0.5. Liquid cultures were grown for an additional 2 hours.
After protein expression, we harvested and lysed the cells with xTractor Lysis Buffer and centrifuged them for sample preparation [5]. This was followed by protein purification through Ni sepharose affinity chromatography, which could isolate our his-tagged SplintR ligase enzymes.
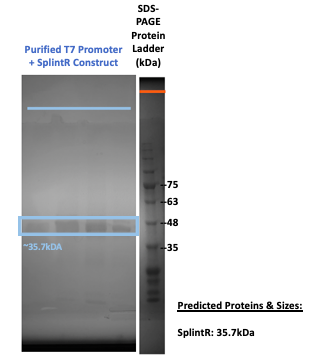
Figure 3: SDS-PAGE on purified proteins with the T7 promoter SplintR ligase expressing construct (BBa_K3352006).
Our results in Figure #3 shows a protein band at around 35.2kDa, which is the molecular weight of our SplintR ligase enzyme (containing the 6x His-tag and GS linker attached to the enzyme). This provides evidence that SplintR ligase was expressed and purified.
Testing whether our purified SplintR ligase works as anticipated, we used it in our Rolling Circle Amplification (RCA) viral diagnostic test. In our diagnostic test, SplintR ligase determines the complementary between the hybrid of DNA padlock probes bound to synthetic viral RNA targets before ligating and circularizing the padlock probe. Using the circularized padlock probe, signal amplification takes place through a pH and color change.
However, because SplintR ligase is stored in unfavorable elution buffer conditions with compounds such as imidazole, we conducted a dialysis buffer exchange. Using Vivaspin® 6 Centrifugal Concentrators, we further enriched our SplintR ligase sample [4]. The new buffer conditions contain 10mM Tris-Hcl, 300mM NaCl, 1mM DTT, 0.1mM EDTA, and 50% glycerol at a pH of 7.4, which is the optimal storage conditions and pH starting point for our RCA test.
Video 1: SARS-CoV-2 RNA RCA Test with Purified SplintR ligase.
Conducting the RCA test on SARS-CoV-2, the PCR tube under letter b is a negative control with no SARS-CoV-2 RNA Target and the PCR tube under letter c is our test group with a SARS-CoV-2 RNA Target. Both tubes use our purified SplintR ligase. The test group yielded a color change from purple to orange in just one hour and ultimately ended at yellow while our negative control remained purple. This results indicates that our SplintR ligase enzymes enabled an accurate detection of the presence of SARS-CoV-2 through its synthetic RNA fragment in our RCA diagnostic test.
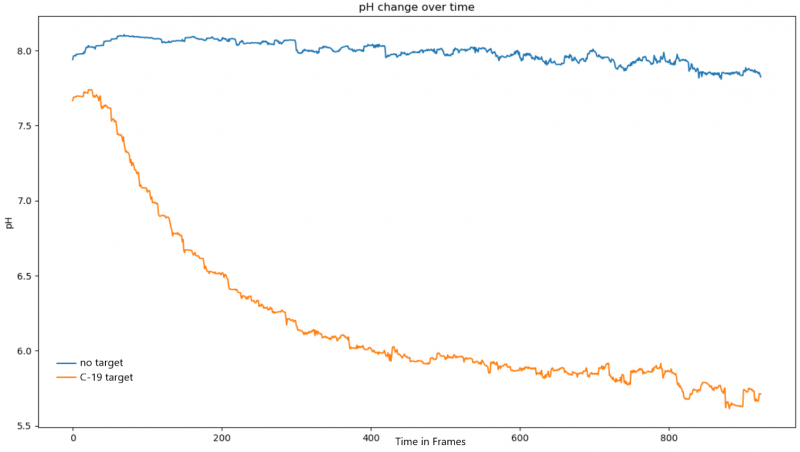
Figure 4: pH over Time Analysis of both tubes in the SARS-CoV-2 RNA RCA Test with Purified SplintR ligase
Using our modeling team’s software to transform our qualitative viral detection readout to quantitative data, we constructed a pH over time graph through the change in hues of both tubes in the video. As shown in Figure #4, there is only minimal fluctuation around the same pH for our negative control and a rapid significant drop in pH of our test group. This is expected due to the production of hydrogen ions from our RCA test that ultimately results in the color change. This provides further validation that SplintR ligase enzyme was expressed, purified, and functioned its intended use.
References
1. A544R - DNA ligase—Paramecium bursaria Chlorella virus 1 (PBCV-1)—A544R gene & protein. (n.d.). Retrieved October 23, 2020, from https://www.uniprot.org/uniprot/O41026
2. Biolabs, N. E. (n.d.). SplintR® ligase | NEB. Retrieved October 20, 2020, from https://international.neb.com/products/m0375-splintr-ligase
3. Krzywkowski, T., & Nilsson, M. (2017). Fidelity of RNA templated end-joining by chlorella virus DNA ligase and a novel iLock assay with improved direct RNA detection accuracy. Nucleic Acids Research, 45(18), e161–e161. https://doi.org/10.1093/nar/gkx708
4. Vivaspin® 6. (n.d.). Retrieved October 23, 2020, from https://www.sartorius.com/shop/ww/en/usd/products-lab-filtration-%26-purification-vivaspin%C2%AE-6-centrifugal-concentrators/c/M_Vivaspin_6
5. XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 835
- 1000COMPATIBLE WITH RFC[1000]
