Difference between revisions of "Part:BBa K3519012"
| (4 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3519012 short</partinfo> | <partinfo>BBa_K3519012 short</partinfo> | ||
| − | This part contains the DNASEI gene downstream to the arabinose inducible | + | This part contains the DNASEI gene (BBa_K3519004) downstream to the arabinose inducible expression system (BBa_K3519010). This is a tightly regulated 'kill switch'. The gene is expressed on the addition of arabinose to the media. Excess glucose in the media represses the expression of the endonuclease thus preventing cell death. All data presented below have been adapted from the literature. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 18: | Line 18: | ||
The AraC protein forms a dimer, each molecule binds to the araI1 and araO2 regulatory regions of the DNA and prevents the binding of RNA polymerase to the P-araBAD promoter region and hence represses the transcription of DNASEI gene. | The AraC protein forms a dimer, each molecule binds to the araI1 and araO2 regulatory regions of the DNA and prevents the binding of RNA polymerase to the P-araBAD promoter region and hence represses the transcription of DNASEI gene. | ||
<br> | <br> | ||
| − | <img src="https://2020.igem.org/wiki/images/0/09/T--IISER-Tirupati_India--partsim2.png" width=" | + | <img src="https://2020.igem.org/wiki/images/0/09/T--IISER-Tirupati_India--partsim2.png" width="800px" hspace="20" vspace="20" style="margin-left:50px"/> |
<br><br> | <br><br> | ||
Figure 2: The ‘inactive’/repressed genetic circuit of the engineered ‘kill switch’. The system here is switched ‘off’ in absence of arabinose in the media. The araC gene is constitutively transcribed in the opposite direction. The AraC dimer binds to the regulatory regions of the promoter and represses the transcription of DNASEI. | Figure 2: The ‘inactive’/repressed genetic circuit of the engineered ‘kill switch’. The system here is switched ‘off’ in absence of arabinose in the media. The araC gene is constitutively transcribed in the opposite direction. The AraC dimer binds to the regulatory regions of the promoter and represses the transcription of DNASEI. | ||
| Line 24: | Line 24: | ||
In the presence of Arabinose (and low amount of glucose sugar), the Arabinose binds to the AraC protein dimer, brings about a conformational change and binds to the regions araI1 and araI2 regions of the DNA releasing the P-araBAD promoter. Now, RNA polymerase binds to the P-araBAD promoter and proceeds with transcription. The incorporation of RNA polymerase at the P-araBAD promoter is enhanced by the CAP-cAMP complex bound to the CAP binding site. As the concentration of cAMP is inversely proportional to the concentration of glucose, for a constant amount of CAP protein, lesser the concentration of glucose, more will be the concentration of cAMP molecules, hence enhanced transcription of DNASEI gene here. | In the presence of Arabinose (and low amount of glucose sugar), the Arabinose binds to the AraC protein dimer, brings about a conformational change and binds to the regions araI1 and araI2 regions of the DNA releasing the P-araBAD promoter. Now, RNA polymerase binds to the P-araBAD promoter and proceeds with transcription. The incorporation of RNA polymerase at the P-araBAD promoter is enhanced by the CAP-cAMP complex bound to the CAP binding site. As the concentration of cAMP is inversely proportional to the concentration of glucose, for a constant amount of CAP protein, lesser the concentration of glucose, more will be the concentration of cAMP molecules, hence enhanced transcription of DNASEI gene here. | ||
<br> | <br> | ||
| − | <img src="https://2020.igem.org/wiki/images/5/5a/T--IISER-Tirupati_India--partsim9.png" width=" | + | <img src="https://2020.igem.org/wiki/images/5/5a/T--IISER-Tirupati_India--partsim9.png" width="800px" hspace="20" vspace="20" style="margin-left:50px"/> |
<br><br> | <br><br> | ||
Figure 3: The ‘active’ genetic circuit of the engineered ‘kill switch’. The system here is switched ‘on’ in addition of arabinose to the media. The araC is constitutively transcribed in the opposite direction. The promoter is activated on recruitment of the cAMP-CAP complex and the arabinose-AraC complex to the regulatory regions in the promoter, leading to increase in the transcription of the DNASEI gene. | Figure 3: The ‘active’ genetic circuit of the engineered ‘kill switch’. The system here is switched ‘on’ in addition of arabinose to the media. The araC is constitutively transcribed in the opposite direction. The promoter is activated on recruitment of the cAMP-CAP complex and the arabinose-AraC complex to the regulatory regions in the promoter, leading to increase in the transcription of the DNASEI gene. | ||
| Line 40: | Line 40: | ||
</html> | </html> | ||
| + | ==Team: BNDS_China 2021, improvements for biosensor== | ||
| + | In this study, we characterized the araBAD promoter and contributed to the iGEM community. | ||
| + | ===Usage in project=== | ||
| + | araBAD is a common promoter that may be present in studies by other iGEM teams, so we wanted to add some data. In our study, we used arabinose to induce the GFP expression and investigated the araBAD promoter. The amount of protein expression was probed by setting a concentration gradient of arabinose and observing the changes in GFP fluorescence intensity. By analyzing the experimental data, we found that the value of GFP/OD showed a logarithmic growth image as the concentration gradient of arabinose increased. | ||
| + | The araBAD promoter is inserted into a constructed plasmid. The arabinose-induced GFP expression plasmid—pGR-ECK120015170—which contains araC, araBAD promoter, RBS, GFP, ECK120015170 terminator and RFP was used to characterize terminator araBAD promoter. The GFP gene is set upstream. | ||
| + | |||
| + | ===Quantitative Characterization=== | ||
| + | Previous studies have shown the feasibility of using green fluorescent protein (GFP) as a quantifiable reporter gene, so it was available to use the intensity of GFP fluorescence to reflect the amount of protein expression upstream. The GFP intensity over OD value can show the amount of protein expression. The ratio due to the different concentration of arabinose is visualized in Figure. | ||
| + | |||
| + | ===Reference=== | ||
| + | 1. Cao, H., & Kuipers, O. P. (2018). Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in bacillus subtilis. Microbial Cell Factories, 17(1). https://doi.org/10.1186/s12934-018-0985-9 | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 14:41, 29 September 2022
araC-araBAD promoter-DNASEI
This part contains the DNASEI gene (BBa_K3519004) downstream to the arabinose inducible expression system (BBa_K3519010). This is a tightly regulated 'kill switch'. The gene is expressed on the addition of arabinose to the media. Excess glucose in the media represses the expression of the endonuclease thus preventing cell death. All data presented below have been adapted from the literature.
Usage and Biology

Figure 1: The ‘native’ genetic circuit of the engineered ‘kill switch’. The araC is constitutively transcribed in the opposite direction.
The AraC protein forms a dimer, each molecule binds to the araI1 and araO2 regulatory regions of the DNA and prevents the binding of RNA polymerase to the P-araBAD promoter region and hence represses the transcription of DNASEI gene.
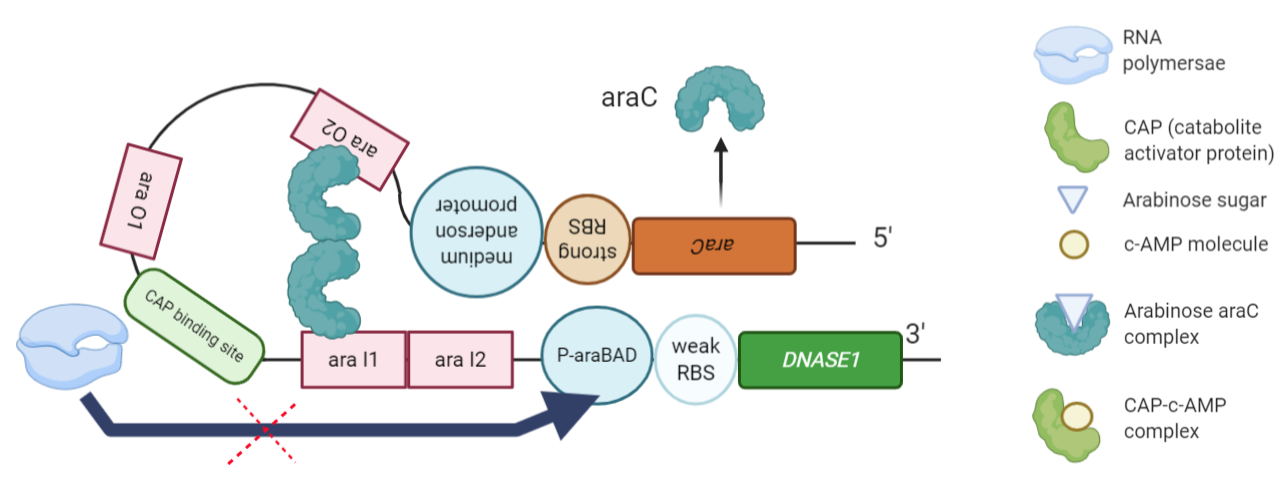
Figure 2: The ‘inactive’/repressed genetic circuit of the engineered ‘kill switch’. The system here is switched ‘off’ in absence of arabinose in the media. The araC gene is constitutively transcribed in the opposite direction. The AraC dimer binds to the regulatory regions of the promoter and represses the transcription of DNASEI.
In the presence of Arabinose (and low amount of glucose sugar), the Arabinose binds to the AraC protein dimer, brings about a conformational change and binds to the regions araI1 and araI2 regions of the DNA releasing the P-araBAD promoter. Now, RNA polymerase binds to the P-araBAD promoter and proceeds with transcription. The incorporation of RNA polymerase at the P-araBAD promoter is enhanced by the CAP-cAMP complex bound to the CAP binding site. As the concentration of cAMP is inversely proportional to the concentration of glucose, for a constant amount of CAP protein, lesser the concentration of glucose, more will be the concentration of cAMP molecules, hence enhanced transcription of DNASEI gene here.
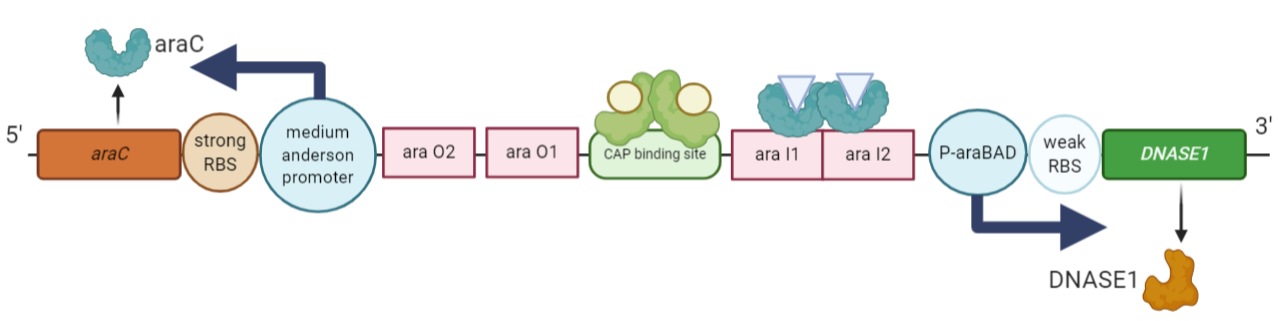
Figure 3: The ‘active’ genetic circuit of the engineered ‘kill switch’. The system here is switched ‘on’ in addition of arabinose to the media. The araC is constitutively transcribed in the opposite direction. The promoter is activated on recruitment of the cAMP-CAP complex and the arabinose-AraC complex to the regulatory regions in the promoter, leading to increase in the transcription of the DNASEI gene.
References:
- Guzman, Luz-Maria, et al. "Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter." Journal of bacteriology 177.14 (1995): 4121-4130
- https://parts.igem.org/Part:BBa_K2442101
- Chen, C. Y., Lu, S. C., & Liao, T. H. (1998). Cloning, sequencing and expression of a cDNA encoding bovine pancreatic deoxyribonuclease I in Escherichia coli: purification and characterization of the recombinant enzyme. Gene, 206(2), 181–184. https://doi.org/10.1016/s0378-1119(97)00582-9
Team: BNDS_China 2021, improvements for biosensor
In this study, we characterized the araBAD promoter and contributed to the iGEM community.
Usage in project
araBAD is a common promoter that may be present in studies by other iGEM teams, so we wanted to add some data. In our study, we used arabinose to induce the GFP expression and investigated the araBAD promoter. The amount of protein expression was probed by setting a concentration gradient of arabinose and observing the changes in GFP fluorescence intensity. By analyzing the experimental data, we found that the value of GFP/OD showed a logarithmic growth image as the concentration gradient of arabinose increased.
The araBAD promoter is inserted into a constructed plasmid. The arabinose-induced GFP expression plasmid—pGR-ECK120015170—which contains araC, araBAD promoter, RBS, GFP, ECK120015170 terminator and RFP was used to characterize terminator araBAD promoter. The GFP gene is set upstream.
Quantitative Characterization
Previous studies have shown the feasibility of using green fluorescent protein (GFP) as a quantifiable reporter gene, so it was available to use the intensity of GFP fluorescence to reflect the amount of protein expression upstream. The GFP intensity over OD value can show the amount of protein expression. The ratio due to the different concentration of arabinose is visualized in Figure.
Reference
1. Cao, H., & Kuipers, O. P. (2018). Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in bacillus subtilis. Microbial Cell Factories, 17(1). https://doi.org/10.1186/s12934-018-0985-9 Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1043
Illegal NheI site found at 1066 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1388
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1223
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1205
