Difference between revisions of "Part:BBa K3610010"
(→Usage and Biology) |
(→Usage and Biology) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | [[File:T--UZurich--CORE ecto.png|300px|thumb|left|Figure 1:CORE (Uniprot Q9SD64)]] | ||
| + | |||
The cold shock protein receptor (CORE) is a plant pattern recognition receptor (PRR) and as such activates host innate immunity through detection of pathogen-associated molecular patterns (PAMPs). | The cold shock protein receptor (CORE) is a plant pattern recognition receptor (PRR) and as such activates host innate immunity through detection of pathogen-associated molecular patterns (PAMPs). | ||
| Line 14: | Line 17: | ||
In our project we use this part to coexpress it together with the ectodomain of BAK1 in S. cerevisiae. The N-terminus of both parts are fused to a split-protein in order to visualize the interaction of the two LRR receptors, used as system for a visual output to confirm dimerization were split-mCherry and split-NanoLuc. | In our project we use this part to coexpress it together with the ectodomain of BAK1 in S. cerevisiae. The N-terminus of both parts are fused to a split-protein in order to visualize the interaction of the two LRR receptors, used as system for a visual output to confirm dimerization were split-mCherry and split-NanoLuc. | ||
| − | + | <br><br><br> | |
==Characterization== | ==Characterization== | ||
| + | |||
| + | ===Expression with YFP=== | ||
| + | |||
| + | All experiments conducted with YFP can be found in more detail on the page for the according construct: [[Part:BBa_K3610046]] | ||
| + | |||
| + | In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed <i>E. coli</i> (DH10alpha) with the plasmids for amplification. | ||
| + | In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same <i>E. coli</i> strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. | ||
| + | For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strengths to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from <i>S. cerevisiae</i>. For termination, this part has the terminator sequence of the enolase 2 protein from <i>S. cerevisiae</i>. | ||
| + | The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in <i>S. cerevisiae</i>. We prepared a medium containing YNB and free amino acids, without tryptophan. | ||
| + | <i>S. cerevisiae</i> cells (AP4) were transfected with the plasmid and then plated on the selective medium. | ||
| + | |||
| + | ====Fluorescence Microscopy==== | ||
| + | |||
| + | After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope. | ||
| + | If expression of YFP (λEx = 515 nm, λEx = 528 nm) can clearly be observed, it is reasonable to assume that the receptor domain is expressed as well, as the YFP is fused to the receptor protein. | ||
| + | Expression of the construct was confirmed. We failed, however, to confirm localization at the cell membrane. | ||
| + | |||
| + | [[File:T--UZurich--Control.png|800px|thumb|none|Figure 2: Confocal microscopy of the normal S. cerevisiae cells (control).]] | ||
| + | [[File:T--UZurich--eCORE.png|800px|thumb|none|Figure 3: Confocal microscopy of S. cerevisiae cells transformed with plasmids containing the COREectodomain fused to YFP. Hardly any increase in fluorescence]] | ||
| + | |||
| + | We also stained the membrane and examined the cells again. | ||
| + | Imaging with a confocal microscope for YFP and the fm4-64 stain shows the spatial overlap of the red fluorescence of the stain and the yellow fluorescence of the protein fused to the receptors. | ||
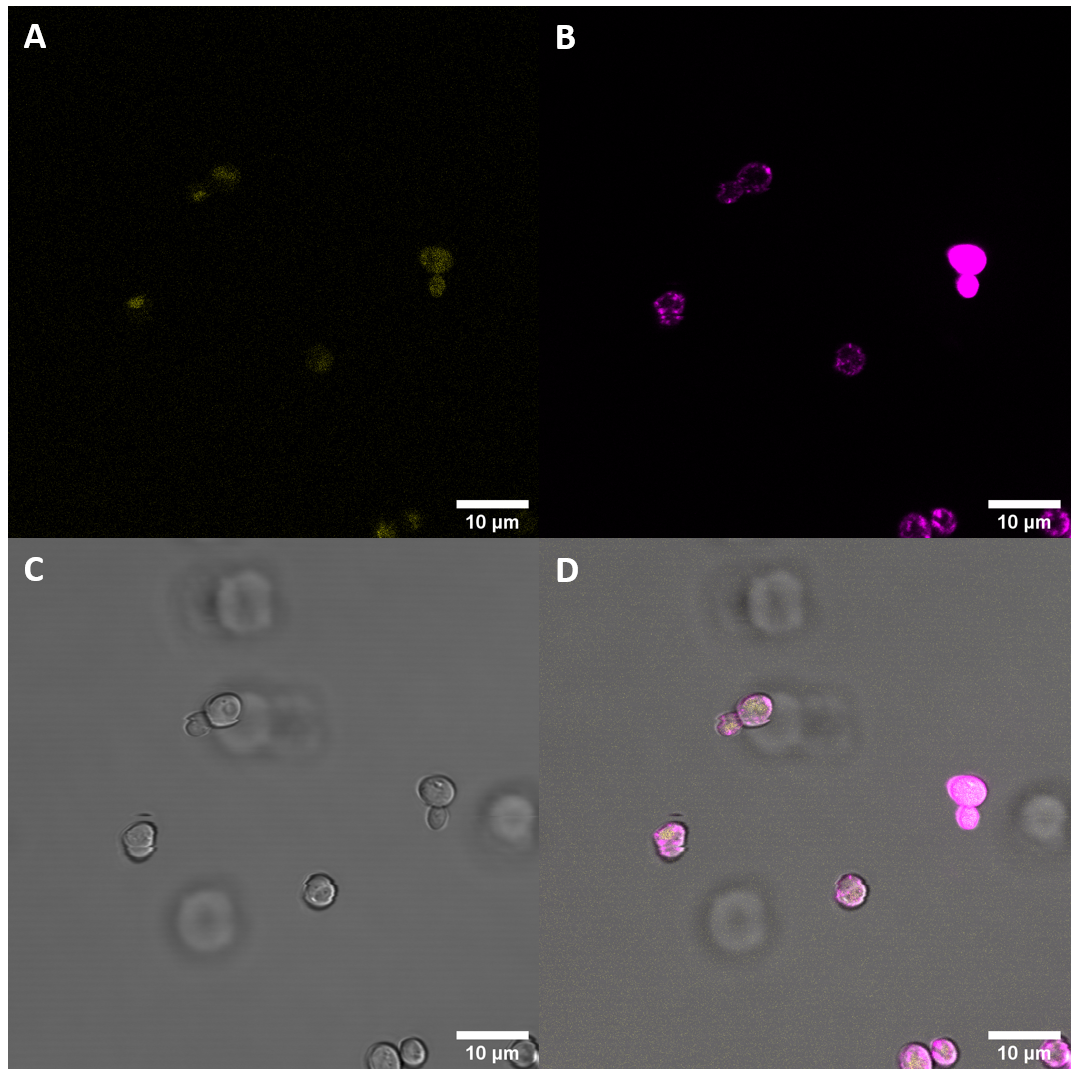
| + | [[File:T--UZurich--UT Membrane Stain.png|440px|thumb|left|Figure 4: Untransformed Control,(A) : YFP, (B) : FM4-64, (C): light field. (D): merge.Imaging of untransfected S. cerevisiae cells reveals hardly any fluorescence within the YFP spectrum]]<br> | ||
| + | [[File:T--UZurich--CORE Membrane Stain.png|440px|thumb|none|Figure 5: eBAK <i>S. cerevisiae</i>: (A) : YFP, (B) : FM4-64, (C) : light field. (D): merge.Most eCORE cells only show weak fluorescence and no localization at the plasma membrane]] | ||
| + | |||
| + | ====Analysis with Plate Reader==== | ||
| + | In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of the CORE receptor with other plant PRRs. | ||
| + | We transformed <i>S. cerevisiae</i> cells with plasmids containing the following receptors (fused to YFP): full length BAK1 with native signal peptide (BAK+), full length BAK1 with alpha-Factor from yeast (BAK-), BAK1 ectodomain (eBAK), EFR ectodomain (eEFR) and the CORE ectodomain (CORE). | ||
| + | |||
| + | For each biological replicate, the optical density at absorbance of 600 nm (OD600) and the fluorescence levels were measured three times. | ||
| + | |||
| + | [[File:T--UZurich--Spectrometer1.png|500px|none|left|Figure 6: Fluorescence values standardized for OD600 of the different receptors (C=Control). Cells with BAK+ showed only weak fluorescence, while BAK-, eBAK and eEFR showed a strong increase in the fluorescence levels. CORE did not display any increase when compared with untreated <i>S. cerevisiae</i> cells (autofluorescence).]] | ||
===Usage with split-NanoLuc=== | ===Usage with split-NanoLuc=== | ||
| + | All experiments conducted with YFP can be found in more detail on the page for the according construct: [[Part:BBa_K3610051]] | ||
| + | |||
We fused this part to the SmallBit part of the NanoBit system, a split luciferase (CORE:SBit). We managed to express the CORE:SBit together with the ectodomain of the plant PRR BAK1 which was fused to the LargeBit part of the split NanoLuc protein (eBAK1:LBit). We were able to express both constructs in <i>S. cerevisiae</i>. For a more detailed description see the parts: [[Part:BBa K3610038]] and [[Part:BBa K3610051]]. | We fused this part to the SmallBit part of the NanoBit system, a split luciferase (CORE:SBit). We managed to express the CORE:SBit together with the ectodomain of the plant PRR BAK1 which was fused to the LargeBit part of the split NanoLuc protein (eBAK1:LBit). We were able to express both constructs in <i>S. cerevisiae</i>. For a more detailed description see the parts: [[Part:BBa K3610038]] and [[Part:BBa K3610051]]. | ||
| Line 37: | Line 74: | ||
The average values of the four measurements are displayed in the plot below. | The average values of the four measurements are displayed in the plot below. | ||
| − | [[File:T--UZurich--NanoLuc CORE.png|600px|thumb|none|Figure | + | [[File:T--UZurich--NanoLuc CORE.png|600px|thumb|none|Figure 7: Average Luminescence over time]] |
eBAK1 coexpressed with eCORE showed an increase in luminescence, although the effect was much smaller when compared with eEFR. The results again suggest, that addition of the bacterial elicitor csp22, which initiates interaciton between CORE and BAK1 does not increase the luminescence levels as samples without csp22 added showed greater luminescence than samples which were treated with this bacterial epitope. | eBAK1 coexpressed with eCORE showed an increase in luminescence, although the effect was much smaller when compared with eEFR. The results again suggest, that addition of the bacterial elicitor csp22, which initiates interaciton between CORE and BAK1 does not increase the luminescence levels as samples without csp22 added showed greater luminescence than samples which were treated with this bacterial epitope. | ||
Latest revision as of 22:10, 27 October 2020
CORE ectodomain from S. lycopersicum
This part compromises the cell surface receptor CORE of S. lycopersicum and contains the ectodomain and the transmembrane domain, including the juxtamembrane domain. It does not, however, include the native signal sequence in order to make it easier to express the CORE ectodomain in different organisms that do not recognize the signal sequence.
Usage and Biology
The cold shock protein receptor (CORE) is a plant pattern recognition receptor (PRR) and as such activates host innate immunity through detection of pathogen-associated molecular patterns (PAMPs).
CORE is a leucine-rich repeat receptor-like kinase with 22 LRRs, there additionally is a 6 amino acid insert at LRR 11. It consists of an extracellular domain that perceives an epitope, csp22, from the highly conserved nucleic acid binding motif RNP-1 of bacterial cold-shock proteins (CSPs), which are highly abundant proteins found in the cytosol of bacteria. Further domains are a single pass transmembrane domain and an intracellular kinase domain (The sequence encoding the kinase domain is not in this part).
Interaction of CORE with brassinosteroid-associated kinase (BAK)1 is necessary for inducing an immune response in the plant. The dimerization of CORE and BAK1 depends on the csp22, the ligand of CORE.
The function of CORE in S. lycopersicum has been confirmed by expressing the receptor in A. thaliana, which made the plant responsive to csp22, a PAMP that is otherwise not perceived by PRRs from A. thaliana.
In our project we use this part to coexpress it together with the ectodomain of BAK1 in S. cerevisiae. The N-terminus of both parts are fused to a split-protein in order to visualize the interaction of the two LRR receptors, used as system for a visual output to confirm dimerization were split-mCherry and split-NanoLuc.
Characterization
Expression with YFP
All experiments conducted with YFP can be found in more detail on the page for the according construct: Part:BBa_K3610046
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strengths to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from S. cerevisiae. For termination, this part has the terminator sequence of the enolase 2 protein from S. cerevisiae. The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in S. cerevisiae. We prepared a medium containing YNB and free amino acids, without tryptophan. S. cerevisiae cells (AP4) were transfected with the plasmid and then plated on the selective medium.
Fluorescence Microscopy
After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope. If expression of YFP (λEx = 515 nm, λEx = 528 nm) can clearly be observed, it is reasonable to assume that the receptor domain is expressed as well, as the YFP is fused to the receptor protein. Expression of the construct was confirmed. We failed, however, to confirm localization at the cell membrane.
We also stained the membrane and examined the cells again. Imaging with a confocal microscope for YFP and the fm4-64 stain shows the spatial overlap of the red fluorescence of the stain and the yellow fluorescence of the protein fused to the receptors.
Analysis with Plate Reader
In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of the CORE receptor with other plant PRRs. We transformed S. cerevisiae cells with plasmids containing the following receptors (fused to YFP): full length BAK1 with native signal peptide (BAK+), full length BAK1 with alpha-Factor from yeast (BAK-), BAK1 ectodomain (eBAK), EFR ectodomain (eEFR) and the CORE ectodomain (CORE).
For each biological replicate, the optical density at absorbance of 600 nm (OD600) and the fluorescence levels were measured three times.
Usage with split-NanoLuc
All experiments conducted with YFP can be found in more detail on the page for the according construct: Part:BBa_K3610051
We fused this part to the SmallBit part of the NanoBit system, a split luciferase (CORE:SBit). We managed to express the CORE:SBit together with the ectodomain of the plant PRR BAK1 which was fused to the LargeBit part of the split NanoLuc protein (eBAK1:LBit). We were able to express both constructs in S. cerevisiae. For a more detailed description see the parts: Part:BBa K3610038 and Part:BBa K3610051.
After transfection of the cells with the two plasmids, we tested whether the split-NanoLuc proteins would be able to interact, which would reconstitute its functionality. We further were interested in the interaction between the CORE and BAK1 ectoomain. CORE and BAK1 naturally interact upon binding of csp22. Should both receptors be expressed properly in the cell membrane of S. cerevisiae, the ligand will be able to bind to the CORE ectodomain. If this is sufficient to drive dimerization of the two receptor in S. cerevisiae, then the NanoLuc parts will be more likely to interact as well, which would lead to a higher amount of functional luciferase proteins in the cell.
To test for dimerization of the luciferase parts and also the two receptors, an assay with a luminometer was performed.
Luminescence assay
Samples with cells which were transfected with the two mentioned plasmids (eBAK1 and eCORE) and samples containing S. cerevisiae cells that were not transfected with any plasmids (UT).
Optical densities (OD600) of all samples were adjusted to 0.34.
For each type of sample, three types of measurements were made:
- 1 µL of deionized water added (no elicitor)
- 1 µL of epitope elf18 added
- 1 µL of epitope csp22 added
Each measurement was done four times with sample size 50µL. To each well 50 µL NanoGlo solution was added (50:1 buffer to furimazine)
The average values of the four measurements are displayed in the plot below.
eBAK1 coexpressed with eCORE showed an increase in luminescence, although the effect was much smaller when compared with eEFR. The results again suggest, that addition of the bacterial elicitor csp22, which initiates interaciton between CORE and BAK1 does not increase the luminescence levels as samples without csp22 added showed greater luminescence than samples which were treated with this bacterial epitope.
These results led us to the conclusion that our plasmids get expressed. It further has been shown that the NanoBit parts fused to the receptors are able to interact and reconstitute their functionality as a funcitonal NanoLuc protein which catalyzes the reaction of furimazine to furimamide, which gives a luminescent output. In our case, however, receptor-specific bacterial epitopes did not increase luminescence levels when the receptors were expressed in S. cerevisiae. It seems that there is no or very little csp22-driven dimerization of the two receptor-ectodomains CORE and BAK1.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1467
Illegal BamHI site found at 100
Illegal BamHI site found at 1492
Illegal BamHI site found at 1844 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]