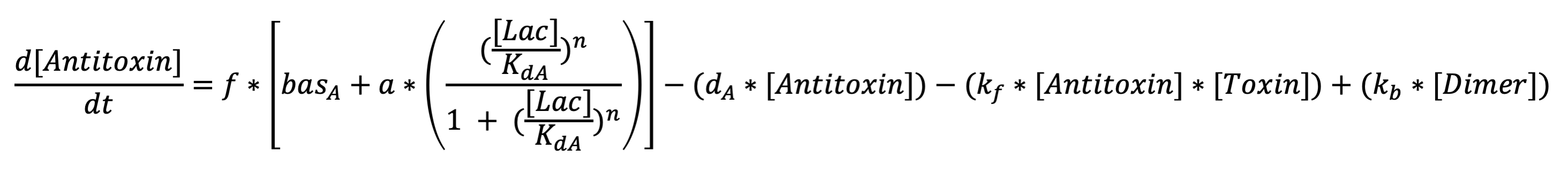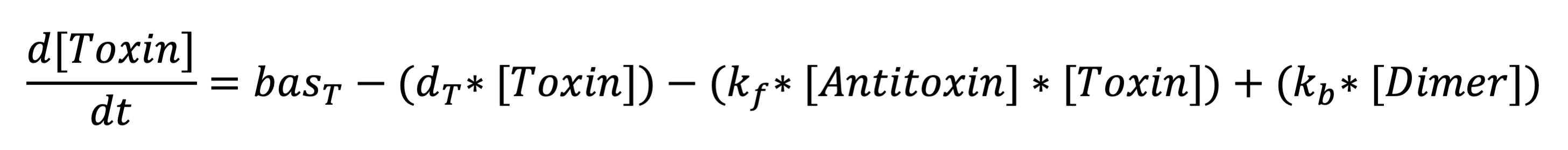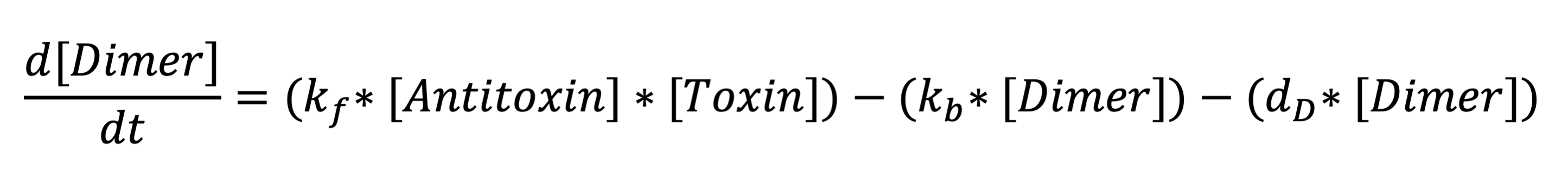Difference between revisions of "Part:BBa K3419005"
Swindemuth (Talk | contribs) (→Modeling) |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 35: | Line 35: | ||
The first equation represents the rate at which antiholin is being produced . The terms respectively represent expression from the lactate promoter, removal due to degradation, removal due to the binding to holin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin-antiholin dimer. | The first equation represents the rate at which antiholin is being produced . The terms respectively represent expression from the lactate promoter, removal due to degradation, removal due to the binding to holin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin-antiholin dimer. | ||
| − | + | Antitoxin differential equation: | |
[[File:Antitoxin.png|700px|center]] | [[File:Antitoxin.png|700px|center]] | ||
| Line 41: | Line 41: | ||
The second equation represents the rate at which holin is being produced. The terms respectively represent the expression from the constitutive promoter, removal due to degradation, removal due to binding to antiholin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin from the holin-antiholin dimer. | The second equation represents the rate at which holin is being produced. The terms respectively represent the expression from the constitutive promoter, removal due to degradation, removal due to binding to antiholin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin from the holin-antiholin dimer. | ||
| − | Toxin | + | Toxin differential equation: |
[[File:Toxin.png|700px|center]] | [[File:Toxin.png|700px|center]] | ||
| Line 47: | Line 47: | ||
The third equation represents the rate at which the holin-antiholin dimer is formed. The terms respectively represent the reaction rate at which holin and antiholin bind, the rate at which the dimer disassociates, and the rate at which the dimer degrades. | The third equation represents the rate at which the holin-antiholin dimer is formed. The terms respectively represent the reaction rate at which holin and antiholin bind, the rate at which the dimer disassociates, and the rate at which the dimer degrades. | ||
| − | Toxin-antitoxin complex | + | Toxin-antitoxin complex differential equation: |
[[File:Dimer.png|700px|center]] | [[File:Dimer.png|700px|center]] | ||
| Line 93: | Line 93: | ||
<td>Concentration of toxin-antitoxin dimer</td> | <td>Concentration of toxin-antitoxin dimer</td> | ||
<td>--</td> | <td>--</td> | ||
| − | <td--</td> | + | <td>--</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>f</td> | <td>f</td> | ||
<td>Fluorescent Scaling Factor</td> | <td>Fluorescent Scaling Factor</td> | ||
| − | <td> = const. | + | <td> = const. expression / (basA + a) = (23.22 uM/min) / (7790 uM/min) = 2.98 *10^-3</td> |
| − | <td>[4][5]</td> | + | <td>[4], [5]</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<tr> | <tr> | ||
<td>bas<sub>A</sub></td> | <td>bas<sub>A</sub></td> | ||
| − | <td>Basal | + | <td>Basal expression of lactate promoter</td> |
<td>479.3 [uM/min]</td> | <td>479.3 [uM/min]</td> | ||
<td>[4]</td> | <td>[4]</td> | ||
| Line 146: | Line 146: | ||
<tr> | <tr> | ||
<td>k<sub>f</sub></td> | <td>k<sub>f</sub></td> | ||
| − | <td>Rate const, forward | + | <td>Rate const, forward reaction: T + A ↔ TA</td> |
<td>1.2 * 10^-4 [uM^-1min^-1]</td> | <td>1.2 * 10^-4 [uM^-1min^-1]</td> | ||
<td>[1] </td> | <td>[1] </td> | ||
| Line 152: | Line 152: | ||
<tr> | <tr> | ||
<td>k<sub>b</sub></td> | <td>k<sub>b</sub></td> | ||
| − | <td>Rate const, reverse | + | <td>Rate const, reverse reaction: T + A ↔ TA</td> |
<td>0.3 * 10^-4 [min^-1]</td> | <td>0.3 * 10^-4 [min^-1]</td> | ||
<td>[4] </td> | <td>[4] </td> | ||
| Line 158: | Line 158: | ||
<tr> | <tr> | ||
<td>bas<sub>T</sub></td> | <td>bas<sub>T</sub></td> | ||
| − | <td>Basal | + | <td>Basal expression of weak constitutive promoter (assume infinite lactate)</td> |
<td>23.22 uM/min for strong promoter J23100, scale by relative strength of J23108 * (1303 / 2547)= 11.87 uM/min= 0.198 [uM/s]</td> | <td>23.22 uM/min for strong promoter J23100, scale by relative strength of J23108 * (1303 / 2547)= 11.87 uM/min= 0.198 [uM/s]</td> | ||
| − | <td>[5][7] </td> | + | <td>[5], [7] </td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
Latest revision as of 01:16, 19 October 2020
Contents
Holin/Anti-Holin Kill Switch with Degradation Tag and Lactate Inducible Promoter
Mechanism
Note: All work on this part was done virtually through literature research due to COVID-19 restrictions.
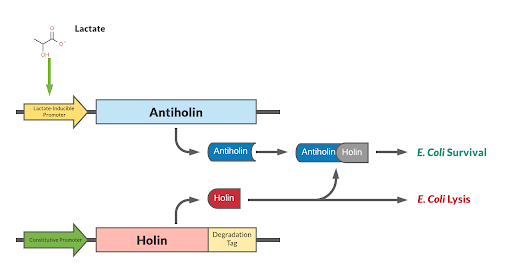
Our kill switch consists of a holin-antiholin toxin-antitoxin system. The holin protein creates holes in the bacterial cell membrane, which allows endolysin protein to pass through and degrade the cell wall, killing the cell. The antiholin protein binds to the holin protein, inactivating it. When both proteins are expressed, whether the cell dies depends on the relative concentrations of both the holin and antiholin components. [1].
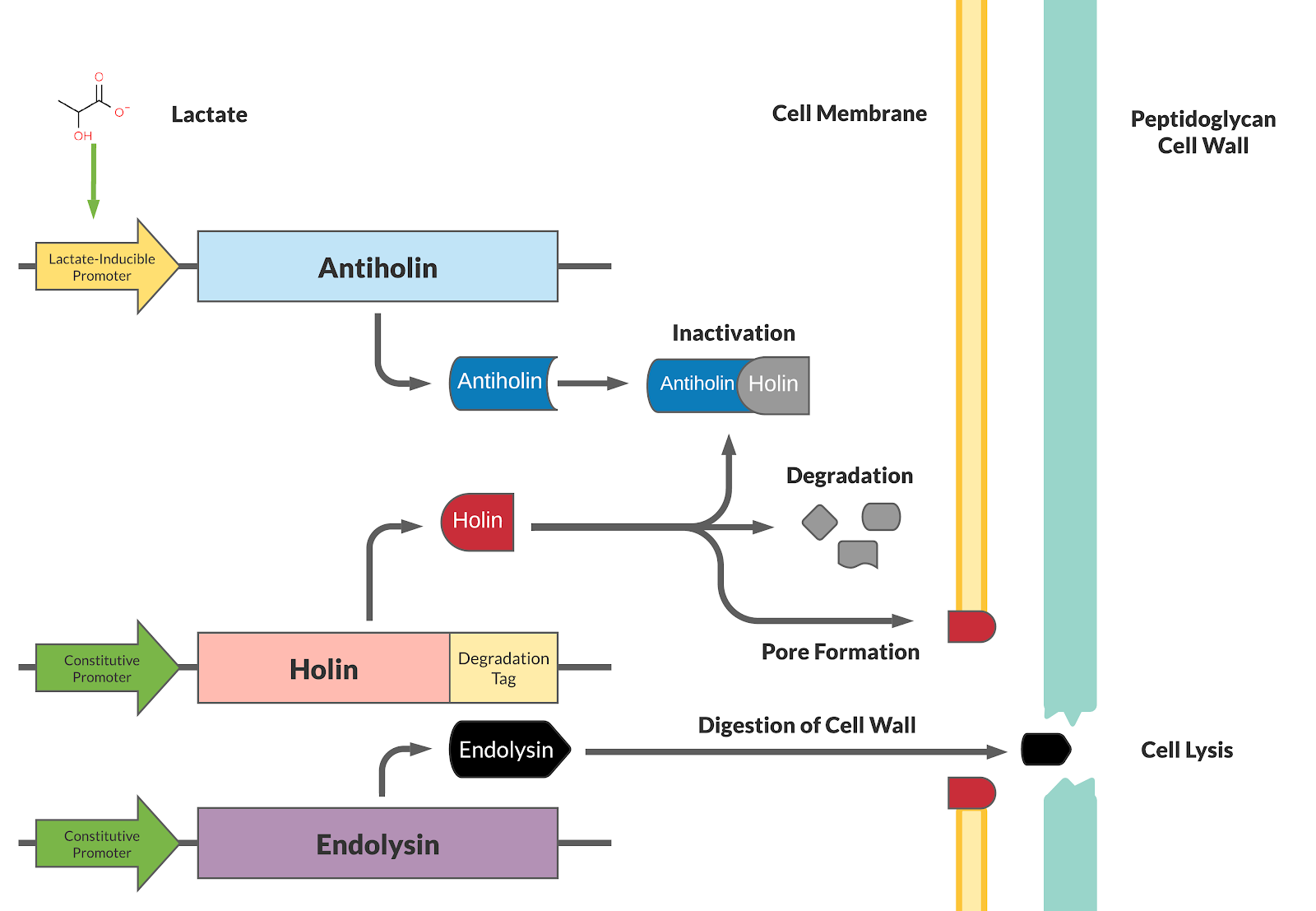
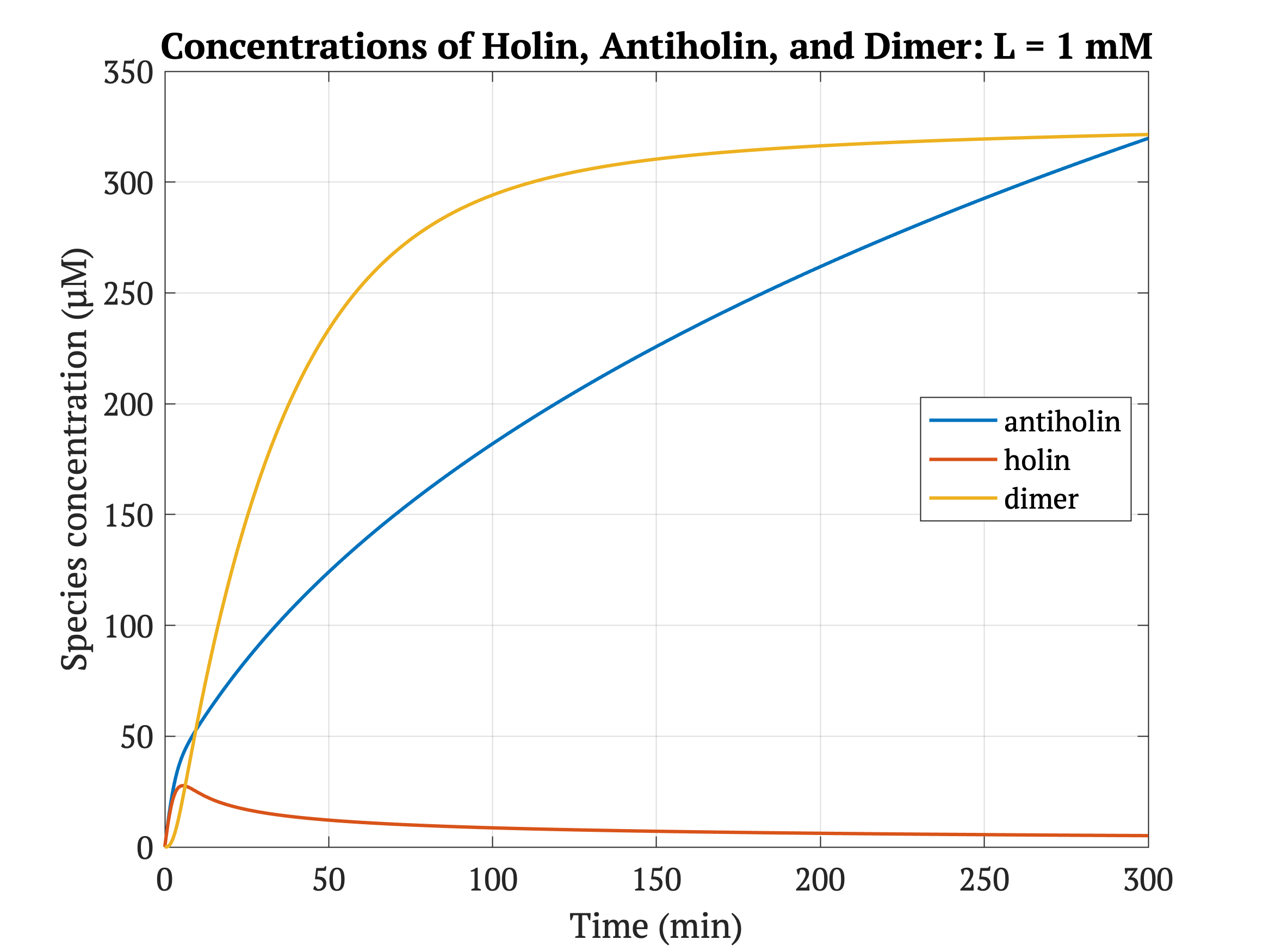
This BioBrick has been designed as a modification to a traditional holin/antiholin kill switch so that the expression of antiholin is controlled by a lactate-inducible promoter, as research shows that lactate concentrations are higher in tumor environments (10-30 mM) compared to healthy tissue (1.5-3 mM) [3]. The toxin, holin, is expressed under a constitutive promoter, as in a typical holin/antiholin kill switch. When in the presence of high lactate, enough antitoxin (antiholin) is produced under the lactate inducible promoter to significantly counteract holin. Otherwise, antiholin is not sufficiently produced, and the bacteria soon die. This allows the bacteria to localize and survive in high-lactate environments such as tumor cells, and die in other environments (i.e. healthy human tissue), as shown in Figure 2.
Improvement
A degradation tag is attached to the holin protein, which marks it for disposal and reduces its half-life in the cell. This limits the activity of the holin toxin prevents its buildup over time, allowing the bacterial population to survive indefinitely as long as it is within a high-lactate tumor environment.
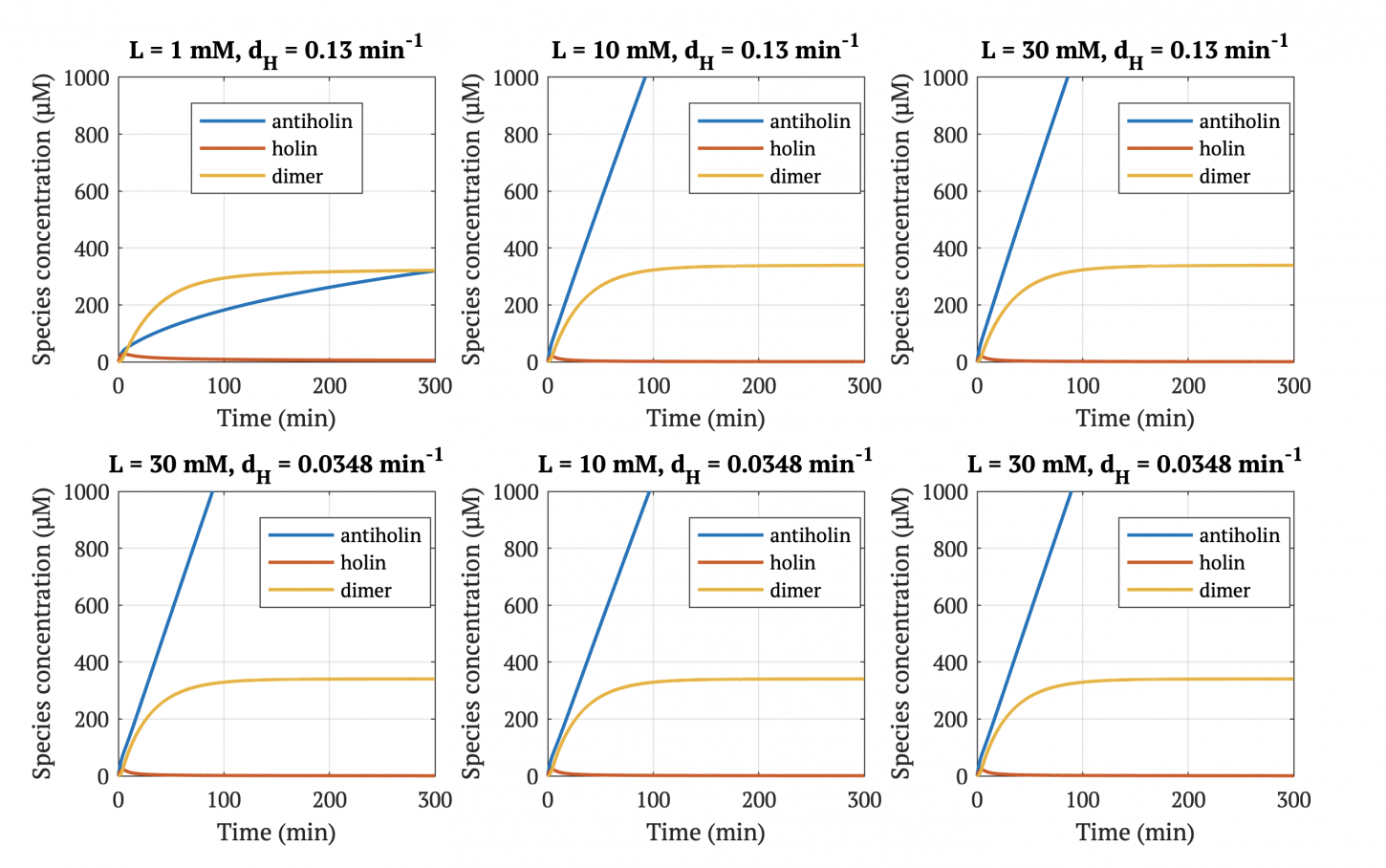
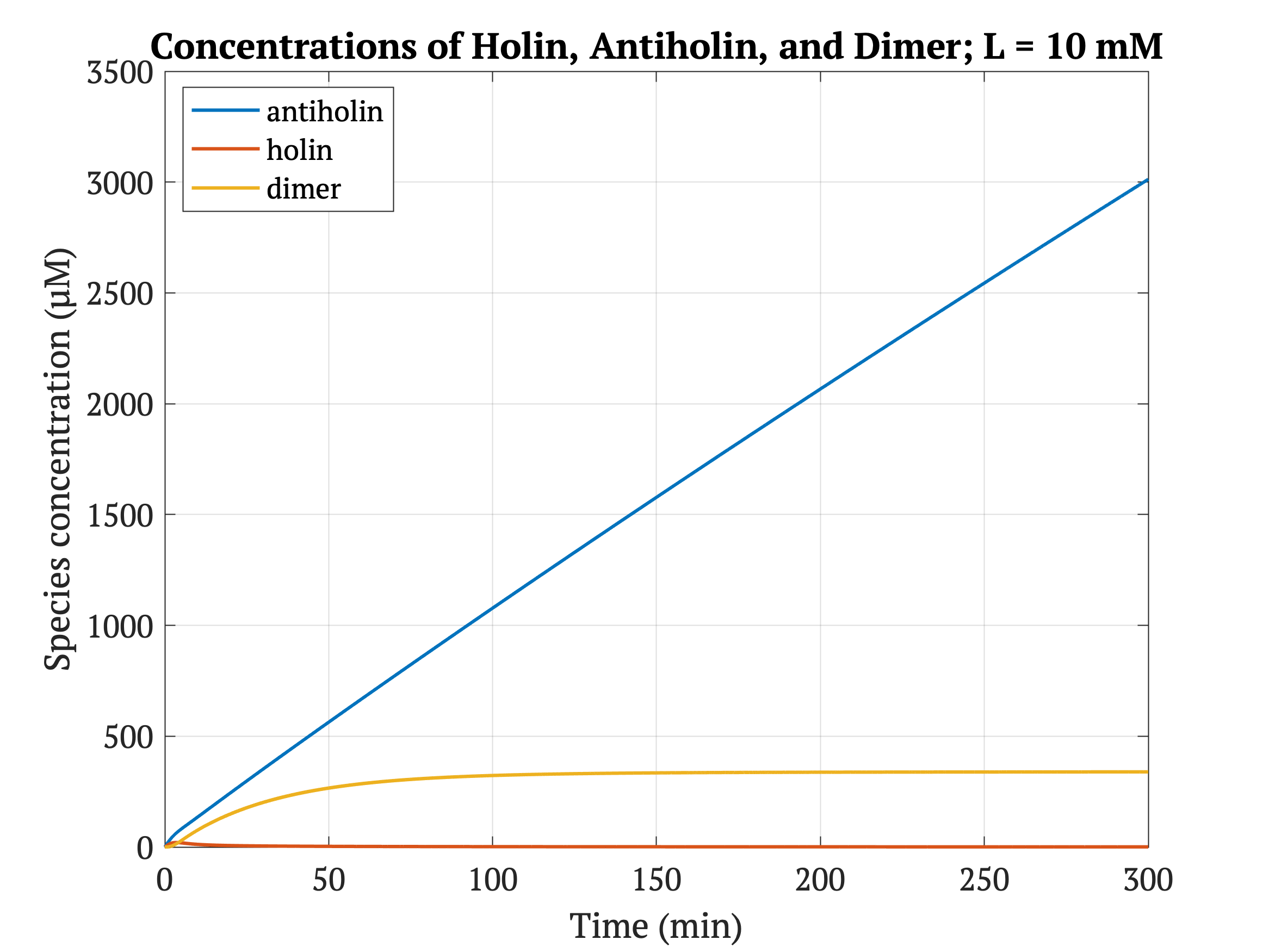
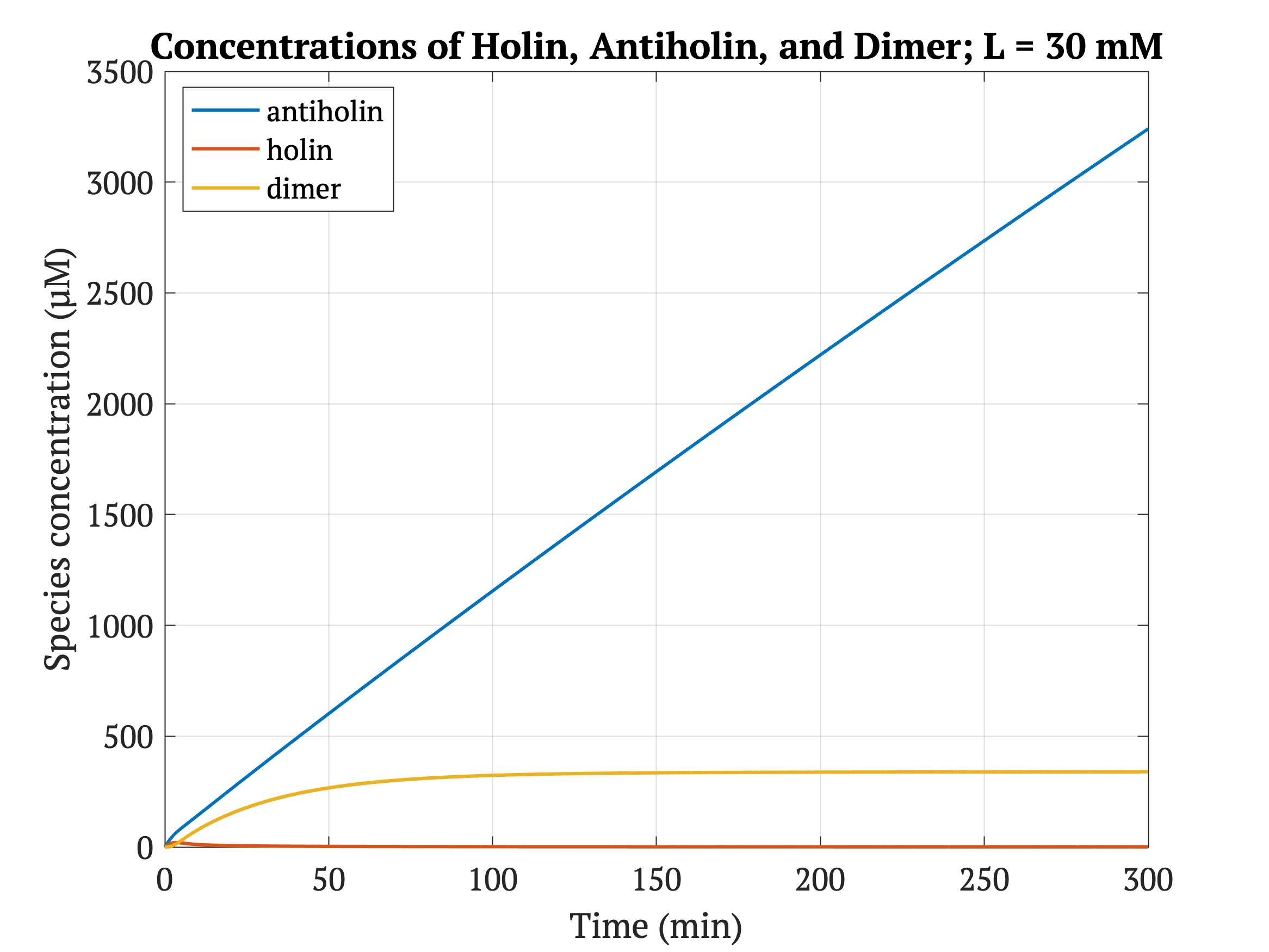
With our improved holin part (ie. with degradation tag), there is good distinction between physiologically normal levels of lactate, and cancerous levels of lactate. In other words, with the degradation tag, the bacteria will survive in a tumor environment, and die in healthy tissue.
However, without our improvement (ie. without degradation tag), as can be seen in the bottom panel of Figure 3, the holin concentration of all of them is between 0.65 uM and 0.75 uM for concentrations throughout 1 mM to 30 mM, both of which would allow survival. Since this range all allows for bacterial survival, this kill switch would not be specific to cancer tissue without the degradation tag.
Modeling
The following three differential equations were used to model our holin-antiholin kill switch. These equations were solved using Matlab’s built-in ordinary differential equation solver ode45.
The first equation represents the rate at which antiholin is being produced . The terms respectively represent expression from the lactate promoter, removal due to degradation, removal due to the binding to holin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin-antiholin dimer.
Antitoxin differential equation:
The second equation represents the rate at which holin is being produced. The terms respectively represent the expression from the constitutive promoter, removal due to degradation, removal due to binding to antiholin (to form the inactivated holin-antiholin dimer), and addition due to disassociation of the holin from the holin-antiholin dimer.
Toxin differential equation:
The third equation represents the rate at which the holin-antiholin dimer is formed. The terms respectively represent the reaction rate at which holin and antiholin bind, the rate at which the dimer disassociates, and the rate at which the dimer degrades.
Toxin-antitoxin complex differential equation:
The MATLAB scripts corresponding to the mCardinal modeling can be downloaded here:
File:ModelingCodeCornelliGEM2020.zip
Table 1. Parameter Values for Kill Switch Modeling
| Variable | Parameter | Value | Source |
|---|---|---|---|
| [Antitoxin] | Concentration of Antitoxin | -- | -- |
| [Toxin] | Concentration of Toxin | -- | -- |
| [Dimer] | Concentration of toxin-antitoxin dimer | -- | -- |
| f | Fluorescent Scaling Factor | = const. expression / (basA + a) = (23.22 uM/min) / (7790 uM/min) = 2.98 *10^-3 | [4], [5] |
| basA | Basal expression of lactate promoter | 479.3 [uM/min] | [4] |
| KdA | Dissociation constant for lactate promoter | 1075 [uM] | [4] |
| n | Exponent | 1.326 | [4] |
| a | Induced transcription scaling factor | 7311 [uM/min] | [4] |
| dA | Rate const, degradation of antitoxin (degradation tag) | 0.13 [min^-1] | [6] |
| dT | Rate const, degradation of toxin | 0.0348 [min^-1] | [1] |
| [Lac] | Lactate concentration | 1 to 30 [mM] | [3] |
| kf | Rate const, forward reaction: T + A ↔ TA | 1.2 * 10^-4 [uM^-1min^-1] | [1] |
| kb | Rate const, reverse reaction: T + A ↔ TA | 0.3 * 10^-4 [min^-1] | [4] |
| basT | Basal expression of weak constitutive promoter (assume infinite lactate) | 23.22 uM/min for strong promoter J23100, scale by relative strength of J23108 * (1303 / 2547)= 11.87 uM/min= 0.198 [uM/s] | [5], [7] |
| dD | Rate const, degradation of toxin-antitoxin dimer | 0.0348 [min^-1] | [1] |
Figures 4-6 show the concentrations of holin, antiholin, and the inactivated holin-antiholin dimer at different levels of lactate. At a lethal concentration of 1.66 μM, holin kills the bacterial cell [1]. Whether holin reaches this level depends on the relative concentration of antiholin, whose expression is influenced by the lactate level of the surrounding environment.
References:
[1] Kill Switch. (2013). Retrieved October 14, 2020, from http://2013.igem.org/KillSwitch
[2] Jie, W. (2014). BBa_K1378032. Retrieved October 13, 2020, from https://parts.igem.org/Part:BBa_K1378032
[3] Cruz-López, K. G., Castro-Muñoz, L. J., Reyes-Hernández, D. O., García-Carrancá, A., & Manzo-Merino, J. (2019). Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Frontiers in Oncology, 9. doi:10.3389/fonc.2019.01143
[4] Fomitcheva, A. (2015). BBa_K1847009. Retrieved October 14, 2020, from https://parts.igem.org/Part:BBa_K1847009
[5] Yildirim, N., & Mackey, M. C. (2003). Feedback Regulation in the Lactose Operon: A Mathematical Modeling Study and Comparison with Experimental Data. Biophysical Journal, 84(5), 2841-2851. doi:10.1016/s0006-3495(03)70013-7
[6] Guan, R. (2013). BBa_K1051208. Retrieved October 14, 2020, from https://parts.igem.org/Part:BBa_K1051208
[7] Anderson, J. (2006). BBa_J23117. Retrieved October 14, 2020, from https://parts.igem.org/Part:BBa_J23117