Difference between revisions of "Part:BBa K3332014"
AnnaTaylor (Talk | contribs) (→Usage) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3332014 short</partinfo> | <partinfo>BBa_K3332014 short</partinfo> | ||
| − | We anchor GRHPR protein onto membranes through AIDA to catalyze the reaction of reducing glyoxalic acid and consuming NADPH. We use | + | We anchor GRHPR protein onto membranes through AIDA to catalyze the reaction of reducing glyoxalic acid and consuming NADPH. We use <partinfo>BBa_K880005</partinfo> to construct the expression system and anchor GRHPR on the surface of ''E.coli''. |
| + | |||
| + | ===Biology=== | ||
| + | |||
| + | AIDA is an anchor protein from ''E. coli'', which has been widely used in cell-surface display. GRHPR, a glyoxylate reductase from human liver, can reduce glyoxylic acid when NADPH is used as cofactor. | ||
| + | |||
| + | GRHPR is fused at C terminal with AIDA so that GRHPR can be displayed on the surface of ''E. coli''.<ref> Rumsby G, Cregeen D P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1999, 1446(3): 383-388.</ref><ref>http://2016.igem.org/Team:TJUSLS_China</ref> | ||
| + | <html> | ||
| + | <figure> | ||
| + | <img src="https://2020.igem.org/wiki/images/8/82/T--XMU-China--XMU-China_2020-GRHPR%E9%94%9A%E5%AE%9A.png" width="45%" style="float:center"> | ||
| + | <figcaption> | ||
| + | <p style="font-size:1rem"> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| + | :'''Fig 1'''. mechanism | ||
| + | |||
| + | ===Usage=== | ||
| + | |||
| + | Here, we used <partinfo>BBa_K880005</partinfo> to construct the expression system and demonstrated the effect of GRHPR-AIDA on the surface of ''E. coli''. We obtained the composite part <partinfo>BBa_K3332059</partinfo> and transformed the constructed plasmid into ''E. coli'' BL21 (DE3) to verify its expression. The positive clones were cultivated. | ||
| + | |||
| + | <html> | ||
| + | <figure> | ||
| + | <img src="https://2020.igem.org/wiki/images/2/21/T--XMU-China--XMU-China_2020-J23100_B0034_grhpr-aidA_B0015.png" width="35%" style="float:center"> | ||
| + | <figcaption> | ||
| + | <p style="font-size:1rem"> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| + | :'''Fig 2'''. Gene circuit of GRHPR-AIDA. | ||
| + | |||
| + | ===Characterization=== | ||
| + | '''1.Identification''' | ||
| + | |||
| + | After receiving the synthesized DNA, restriction digestion was done to certify that the plasmid was correct, and the experimental results were shown in figure 3. | ||
| + | <html> | ||
| + | <figure> | ||
| + | <img src="https://2020.igem.org/wiki/images/2/20/T--XMU-China--09031.png" width="60%" style="float:center"> | ||
| + | <figcaption> | ||
| + | <p style="font-size:1rem"> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | :'''Fig 3'''.DNA gel electrophoresis of restriction digest products of GRHPR-AIDA-pSB1C3 (''Xba''l I & ''Pst'' I sites) | ||
| + | |||
| + | '''2.Ability of consuming NADPH''' | ||
| + | |||
| + | We mixed glyoxylic acid solution, NADPH solution and bacteria solution carrying GRHPR-AIDA. Then, we immediately measured OD<sub>340</sub> changes. TECAN<sup>®</sup> Infinite M200 Pro was used to detect OD<sub>340</sub> . And when NADPH is consumed, OD<sub>340</sub> declines. | ||
| + | |||
| + | We successfully got OD340-Time curves of GRHPR fused with 4 types of anchor protein. When using bacteria carrying GRHPR-AIDA, we could see OD<sub>340</sub> decreased as the reaction went on. However, by using bacteria carrying J23100-RBS (<partinfo>BBa_K880005</partinfo>) and GRHPR-Histag as control, we could also find that the slopes of these three curves are similar. The results show that our fusion protein GRHPR-AIDA do not work well. The result is shown in figure 4. | ||
| + | |||
| + | <html> | ||
| + | <figure> | ||
| + | <img src="https://2020.igem.org/wiki/images/a/a5/T--XMU-China--XMU-China_2020-GRHPR%E9%94%9A%E5%AE%9A%E9%85%B6%E6%B4%BB.png" width="40%" style="float:center"> | ||
| + | <figcaption> | ||
| + | <p style="font-size:1rem"> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | :'''Fig 4.''' OD<sub>340</sub>-Time curves of GRHPR fused with 4 types of anchor protein | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | <references/> | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
Latest revision as of 22:58, 27 October 2020
GRHPR-AIDA
We anchor GRHPR protein onto membranes through AIDA to catalyze the reaction of reducing glyoxalic acid and consuming NADPH. We use BBa_K880005 to construct the expression system and anchor GRHPR on the surface of E.coli.
Biology
AIDA is an anchor protein from E. coli, which has been widely used in cell-surface display. GRHPR, a glyoxylate reductase from human liver, can reduce glyoxylic acid when NADPH is used as cofactor.
GRHPR is fused at C terminal with AIDA so that GRHPR can be displayed on the surface of E. coli.[1][2]
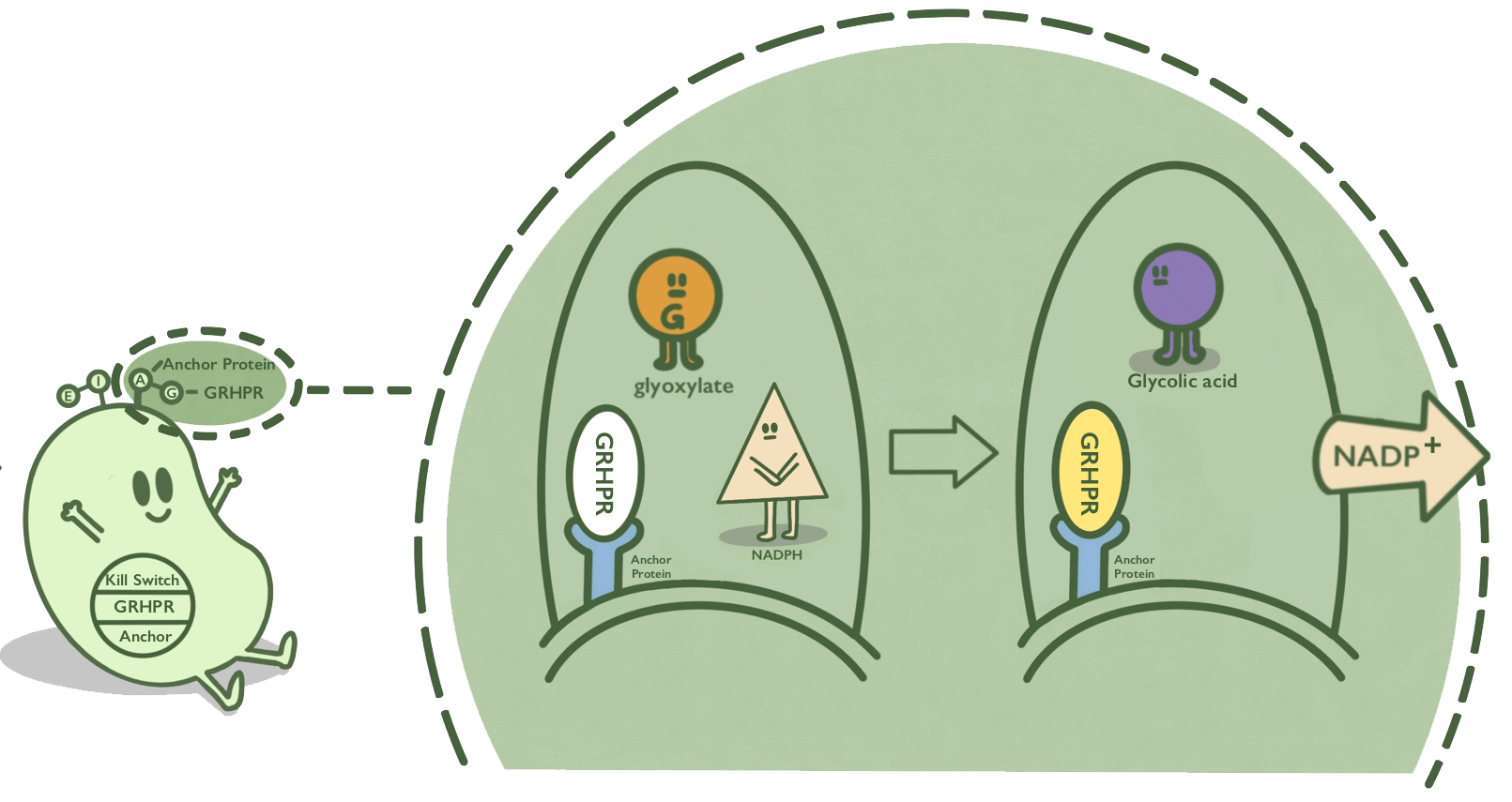
- Fig 1. mechanism
Usage
Here, we used BBa_K880005 to construct the expression system and demonstrated the effect of GRHPR-AIDA on the surface of E. coli. We obtained the composite part BBa_K3332059 and transformed the constructed plasmid into E. coli BL21 (DE3) to verify its expression. The positive clones were cultivated.

- Fig 2. Gene circuit of GRHPR-AIDA.
Characterization
1.Identification
After receiving the synthesized DNA, restriction digestion was done to certify that the plasmid was correct, and the experimental results were shown in figure 3.

- Fig 3.DNA gel electrophoresis of restriction digest products of GRHPR-AIDA-pSB1C3 (Xbal I & Pst I sites)
2.Ability of consuming NADPH
We mixed glyoxylic acid solution, NADPH solution and bacteria solution carrying GRHPR-AIDA. Then, we immediately measured OD340 changes. TECAN® Infinite M200 Pro was used to detect OD340 . And when NADPH is consumed, OD340 declines.
We successfully got OD340-Time curves of GRHPR fused with 4 types of anchor protein. When using bacteria carrying GRHPR-AIDA, we could see OD340 decreased as the reaction went on. However, by using bacteria carrying J23100-RBS (BBa_K880005) and GRHPR-Histag as control, we could also find that the slopes of these three curves are similar. The results show that our fusion protein GRHPR-AIDA do not work well. The result is shown in figure 4.

- Fig 4. OD340-Time curves of GRHPR fused with 4 types of anchor protein
References
- ↑ Rumsby G, Cregeen D P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1999, 1446(3): 383-388.
- ↑ http://2016.igem.org/Team:TJUSLS_China
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2264
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 547
- 1000COMPATIBLE WITH RFC[1000]
