Difference between revisions of "Part:BBa K3593007"
| Line 4: | Line 4: | ||
The aptamer we selected by magnetic beads SELEX for β-amatoxin. The final pool consists of multiple of it which suggests its strong affinity with β-aptamer, above chance. Its binding property is further quantified by ELONA, in which there's a significantly higher binding to beta-BSA conjugate compared with BSA | The aptamer we selected by magnetic beads SELEX for β-amatoxin. The final pool consists of multiple of it which suggests its strong affinity with β-aptamer, above chance. Its binding property is further quantified by ELONA, in which there's a significantly higher binding to beta-BSA conjugate compared with BSA | ||
| + | =Background= | ||
| + | Amatoxins are chemicals present inside the genus Amanita and caused about 90% of mushroom poisoning. Being able to detect it before eating or in the field could possibly make a great decrease in people and animals who died because of poisonous mushrooms. Also being able to detect it in hospital can greatly help doctors in mushroom areas get correct information and do effective diagnosis to save the patient.<br> | ||
| + | Aptamers are oligonucleotides that form secondary structures, giving them the ability to bind targeted molecules, including ions or small molecules, and, in our case, amanitin. BBa_K3593007 is an aptamer for β-amanitin. | ||
| + | =Sequence and features= | ||
| + | <partinfo>BBa_K3593007 SequenceAndFeatures</partinfo> | ||
| + | =Characterisation of this part= | ||
| + | ==Design== | ||
| + | ==Experiment data== | ||
| + | The resultant product of each round of SELEX is about 80bp, as we expected, and the band is bright. As the SELEX progresses, the band seems to be broader, indicating a nonspecific amplification, which might be owing to the increase in the concentration of aptamer with the nonspecific binding site of primer, due to its affinity with the toxin on beads. We thus increase the revealing temperature and the band becomes sharp again. At the last round(round 15), we used all beaming elucidate from round 14 to prepare 2 samples of aptamer pool, 15-1 & 15-2, to ensure an unbiased resultant aptamer pool.<br> | ||
| + | |||
| + | Terminating the process, we found out that the sequence of the aptamer is too short (80bp) to be sequenced. To overcome this dilemma, we purify the PCR product from round 15 and attempts to insert that ssDNA into plasmid PSB1C3 by Gibson assembly.<br> | ||
| + | |||
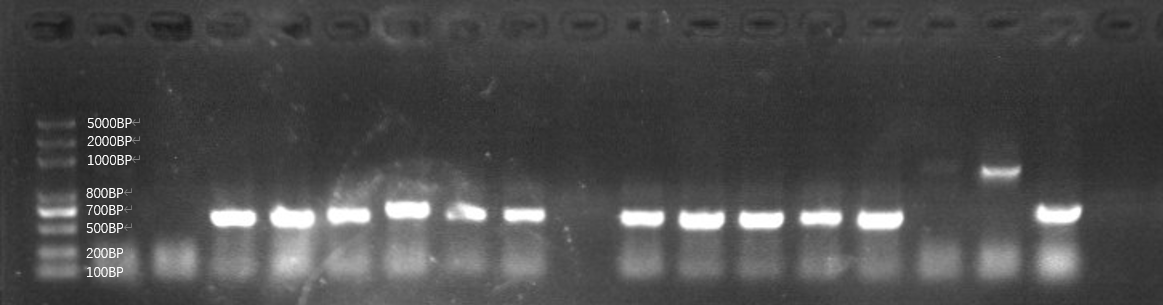
| + | [[File:T--GreatBay_SCIE--Fig.1 Colony PCR of our bacteria that carries the sequence of the aptamer through Gibson Assembly.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.1 Colony PCR of our bacteria that carries the sequence of the aptamer through Gibson Assembly.</center></b></p> | ||
| + | However, as we can tell from the graph, the plasmid digestion is not effective so that sample in lane 15 and 16 might be the template plasmid directly transferred to bacteria DH5α. Then, we also found out even those ones with seemingly correct length have an incorrect sequencing result, indicating that the 80bp aptamer is not assembled to the plasmid and the length is the pure plasmid part being amplified. We attribute this failure as a low assembling efficiency of Gibson assembly specific to a short DNA sequence, hence we assemble the plasmid and aptamer by an alternative method, golden gate.<br> | ||
| + | |||
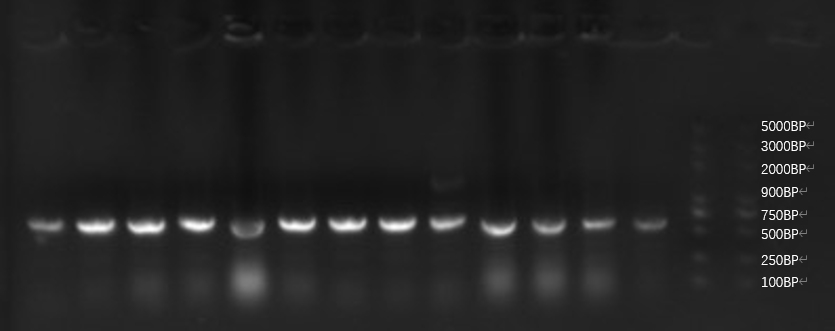
| + | [[File:T--GreatBay_SCIE--Fig.2 Colony PCR of our bacteria that carries the sequence of the aptamer through Golden Gate.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.2 Colony PCR of our bacteria that carries the sequence of the aptamer through Golden Gate.</center></b></p> | ||
| + | All of our sample from golden gate assembly is in the correct length, and the sequencing result indicates successful assembly for all. Here we put our insight into the sequencing result and it's inspiring that one single sequence occurs repetitively among 15 samples of bacteria PCR, 5 times. This implies a possible high concentration of the sequence after PCR, indicating a possibly high affinity to the toxin during fishing.<br> | ||
| + | |||
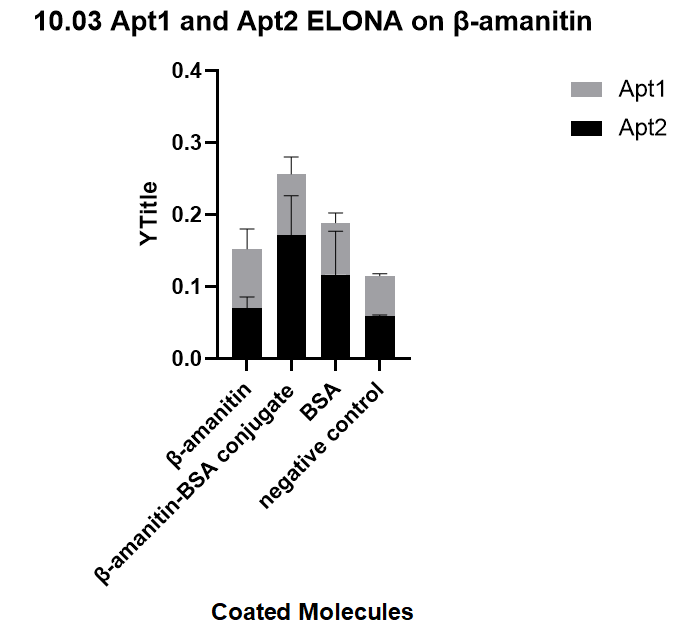
| + | We ordered the aptamers from DNA synthesis companies based on the sequences we obtained from sequencing and verify the specificity of each aptamer towards three targets: beta-amanitin, beta-amanitin-BSA conjugate, and BSA, through ELONA, an experiment that comes from a previous literature[1], with a similar principle to ELISA. This one's binding affinity is successfully proved, and is one of our successfully selected aptamer.<br> | ||
| + | |||
| + | [[File:T--GreatBay_SCIE--Fig.3 ELONA on aptamers selected on our own.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.3 ELONA on aptamers selected on our own.</center></b></p> | ||
| + | =References= | ||
| + | 1. Han Q, Xia X, Jing L, et al. Selection and characterization of DNA aptamer specially targeting α-amanitin in wild mushrooms. SDRP J Food Sci Technol. 2018;3(6):497-508. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 9: | Line 35: | ||
<!-- --> | <!-- --> | ||
| − | + | ||
| − | + | ||
Latest revision as of 05:15, 27 October 2020
ssDNA, aptamer for β-amanitin
The aptamer we selected by magnetic beads SELEX for β-amatoxin. The final pool consists of multiple of it which suggests its strong affinity with β-aptamer, above chance. Its binding property is further quantified by ELONA, in which there's a significantly higher binding to beta-BSA conjugate compared with BSA
Background
Amatoxins are chemicals present inside the genus Amanita and caused about 90% of mushroom poisoning. Being able to detect it before eating or in the field could possibly make a great decrease in people and animals who died because of poisonous mushrooms. Also being able to detect it in hospital can greatly help doctors in mushroom areas get correct information and do effective diagnosis to save the patient.
Aptamers are oligonucleotides that form secondary structures, giving them the ability to bind targeted molecules, including ions or small molecules, and, in our case, amanitin. BBa_K3593007 is an aptamer for β-amanitin.
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterisation of this part
Design
Experiment data
The resultant product of each round of SELEX is about 80bp, as we expected, and the band is bright. As the SELEX progresses, the band seems to be broader, indicating a nonspecific amplification, which might be owing to the increase in the concentration of aptamer with the nonspecific binding site of primer, due to its affinity with the toxin on beads. We thus increase the revealing temperature and the band becomes sharp again. At the last round(round 15), we used all beaming elucidate from round 14 to prepare 2 samples of aptamer pool, 15-1 & 15-2, to ensure an unbiased resultant aptamer pool.
Terminating the process, we found out that the sequence of the aptamer is too short (80bp) to be sequenced. To overcome this dilemma, we purify the PCR product from round 15 and attempts to insert that ssDNA into plasmid PSB1C3 by Gibson assembly.
However, as we can tell from the graph, the plasmid digestion is not effective so that sample in lane 15 and 16 might be the template plasmid directly transferred to bacteria DH5α. Then, we also found out even those ones with seemingly correct length have an incorrect sequencing result, indicating that the 80bp aptamer is not assembled to the plasmid and the length is the pure plasmid part being amplified. We attribute this failure as a low assembling efficiency of Gibson assembly specific to a short DNA sequence, hence we assemble the plasmid and aptamer by an alternative method, golden gate.
All of our sample from golden gate assembly is in the correct length, and the sequencing result indicates successful assembly for all. Here we put our insight into the sequencing result and it's inspiring that one single sequence occurs repetitively among 15 samples of bacteria PCR, 5 times. This implies a possible high concentration of the sequence after PCR, indicating a possibly high affinity to the toxin during fishing.
We ordered the aptamers from DNA synthesis companies based on the sequences we obtained from sequencing and verify the specificity of each aptamer towards three targets: beta-amanitin, beta-amanitin-BSA conjugate, and BSA, through ELONA, an experiment that comes from a previous literature[1], with a similar principle to ELISA. This one's binding affinity is successfully proved, and is one of our successfully selected aptamer.
References
1. Han Q, Xia X, Jing L, et al. Selection and characterization of DNA aptamer specially targeting α-amanitin in wild mushrooms. SDRP J Food Sci Technol. 2018;3(6):497-508.