Difference between revisions of "Part:BBa K3457056"
| (One intermediate revision by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3457056 short</partinfo> | <partinfo>BBa_K3457056 short</partinfo> | ||
| − | a | + | <p> This biological part, given by iGEM Team NEFU-China 2020, is the expression sequence of Green Fluorescent Protein, also called GFP. We use it in the experiment to test arabinose sensor. It also used as the control group in some of our freeze-drying experiments.</p> |
| + | |||
| + | ===Contribution=== | ||
| + | <h3><b>Group: QHFZ-China iGEM 2020</b></h3> | ||
| + | <h3><b>Author: Yixian Yang</b></h3> | ||
| + | <p> TDPs (Tardigrade intrinsically Disordered Proteins) show protective effect to bacteria. However, ralated studies are all use T7 promoter and <i>E. coli</i> BL21 (DE3) strain. This year, we used the Inducible pBad/araC promoter to express a TDP, CAHS 106094, in another starin, when this part was used as the control and reporter. For all the experiments below, we use <i>E. coli</i> DH5α strain.</p> | ||
| + | <h3><b>Design</b></h3> | ||
| + | [[File:T--QHFZ-China--ara-111.png|400px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct.]] | ||
| + | <h3 style="clear:left;"><b>Documentation:</b></h3> | ||
| + | <h4 style="clear:left;"><b>Protocol: </b></h4> | ||
| + | <p style="clear:left;"> The protocol is as below: <br> | ||
| + | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to | ||
| + | grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br> | ||
| + | (2) Add inducer into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 | ||
| + | to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br> | ||
| + | (3) 100 μL bacteria solution solution was put into a well of a 96-well palte. The GFP fluorescence and OD<sub>600</sub> were detected | ||
| + | by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50. | ||
| + | <br> | ||
| + | (4) The value of LB medium was deducted from the result above. GFP / OD<sub>600</sub> was calculated.<br> | ||
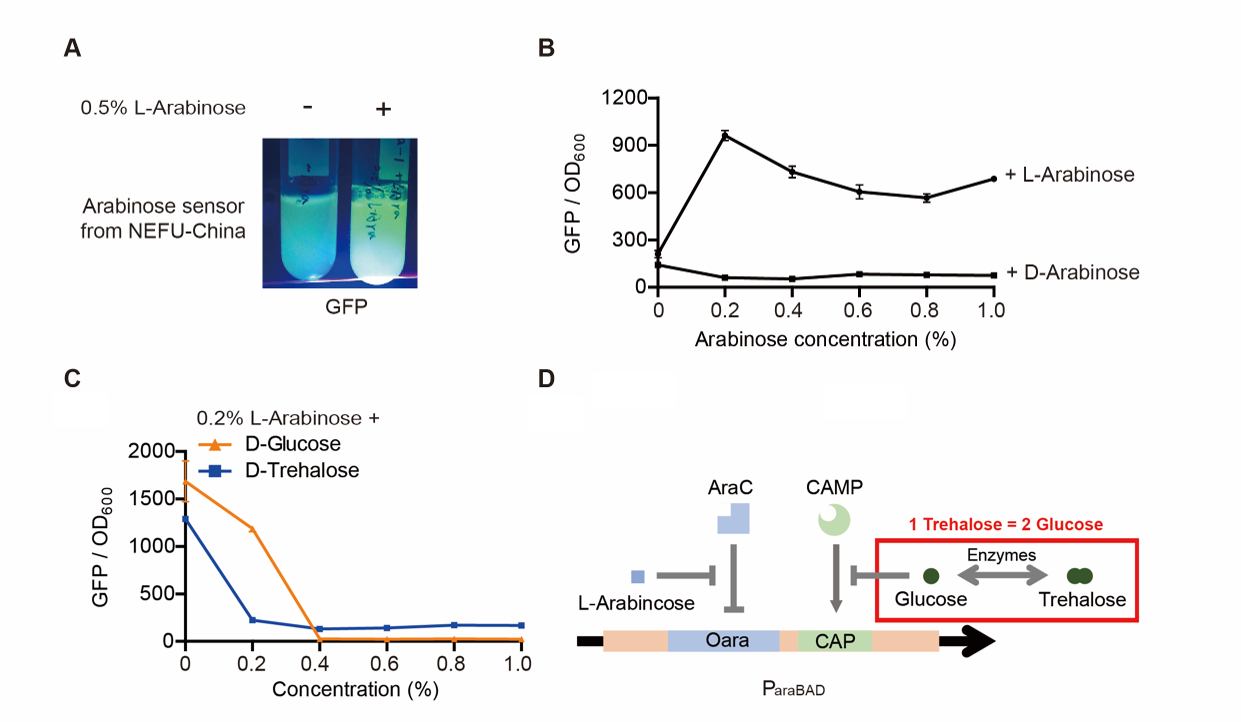
| + | Note: For step (3), The inducer usually was 0.2% L-arabinose. However, it can be changed if we want to study the concentration of the inducer and the effect of D-arabinose. To study whether glucose and trehalose affect the promoter, we even added D-glucose or D-trehalose of different concentrations with 0.2% L-arabinose into the medium. | ||
| + | </p> | ||
| + | <h4>Result</h4> | ||
| + | [[File:T--QHFZ-China--ara-2.png|600px|thumb|left|Figure 2. GFP can be induced by 0.2% L-arabinose, but not D-arabinose. Glucose and trehalose afected the promoter.]] | ||
| + | <p style="clear:left;"> The result is quite clear. First, L-arabinose efficiently induced the expression of GFP. 0.2% arabinose seemed enough. However, D-arabinose did not have any effect. Secondly, glucose and trehalose supressed the promoter, even though 0.2% L-arabinose was added. At the concentration of 0.3%, the effect of trehalose was even wore than that of glucose. The effect of glucose is well-known. However, that of trehalose is not. We give a hypothesis here. One trehalose can be hydrolyzed into two glucose. So 0.3% trehalose equals to 0.6% glucose. Thus, at a low concentration, trehalose is more unsufferable to the promoter than glucose.</p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 02:55, 27 October 2020
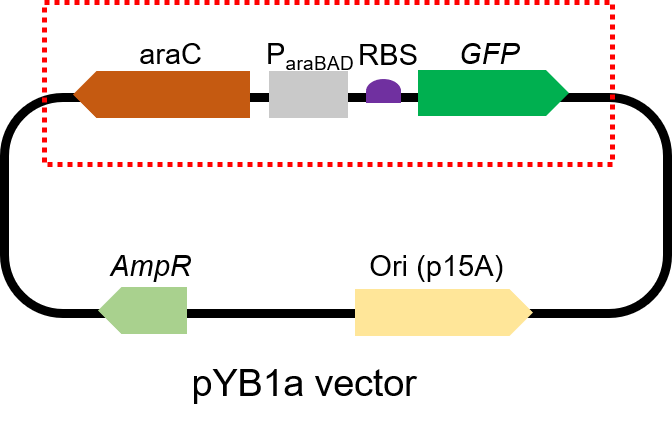
Arac-Pbad-GFP
This biological part, given by iGEM Team NEFU-China 2020, is the expression sequence of Green Fluorescent Protein, also called GFP. We use it in the experiment to test arabinose sensor. It also used as the control group in some of our freeze-drying experiments.
Contribution
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
TDPs (Tardigrade intrinsically Disordered Proteins) show protective effect to bacteria. However, ralated studies are all use T7 promoter and E. coli BL21 (DE3) strain. This year, we used the Inducible pBad/araC promoter to express a TDP, CAHS 106094, in another starin, when this part was used as the control and reporter. For all the experiments below, we use E. coli DH5α strain.
Design
Documentation:
Protocol:
The protocol is as below:
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add inducer into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
(3) 100 μL bacteria solution solution was put into a well of a 96-well palte. The GFP fluorescence and OD600 were detected
by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50.
(4) The value of LB medium was deducted from the result above. GFP / OD600 was calculated.
Note: For step (3), The inducer usually was 0.2% L-arabinose. However, it can be changed if we want to study the concentration of the inducer and the effect of D-arabinose. To study whether glucose and trehalose affect the promoter, we even added D-glucose or D-trehalose of different concentrations with 0.2% L-arabinose into the medium.
Result
The result is quite clear. First, L-arabinose efficiently induced the expression of GFP. 0.2% arabinose seemed enough. However, D-arabinose did not have any effect. Secondly, glucose and trehalose supressed the promoter, even though 0.2% L-arabinose was added. At the concentration of 0.3%, the effect of trehalose was even wore than that of glucose. The effect of glucose is well-known. However, that of trehalose is not. We give a hypothesis here. One trehalose can be hydrolyzed into two glucose. So 0.3% trehalose equals to 0.6% glucose. Thus, at a low concentration, trehalose is more unsufferable to the promoter than glucose.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 1704
Illegal EcoRI site found at 2016
Illegal SpeI site found at 2007
Illegal PstI site found at 2034 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1704
Illegal EcoRI site found at 2016
Illegal SpeI site found at 2007
Illegal PstI site found at 2034 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1704
Illegal EcoRI site found at 2016
Illegal BglII site found at 1998
Illegal BamHI site found at 1144
Illegal BamHI site found at 1913
Illegal XhoI site found at 1281
Illegal XhoI site found at 1989 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 1704
Illegal EcoRI site found at 2016
Illegal SpeI site found at 2007
Illegal PstI site found at 2034 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 1704
Illegal EcoRI site found at 2016
Illegal SpeI site found at 2007
Illegal PstI site found at 2034
Illegal AgeI site found at 979 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961