Difference between revisions of "Part:BBa K3351000"
| (9 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3351000 short</partinfo> | <partinfo>BBa_K3351000 short</partinfo> | ||
| + | ===Summary=== | ||
| + | It is a strong promoter from T7 bacteriophage that can specifically react to T7RNA polymerase. It is a sequence that initiates the transcription of T7 bacteriophage gene. | ||
| + | |||
| + | ===Characterization by NWU-CHINA-A 2021=== | ||
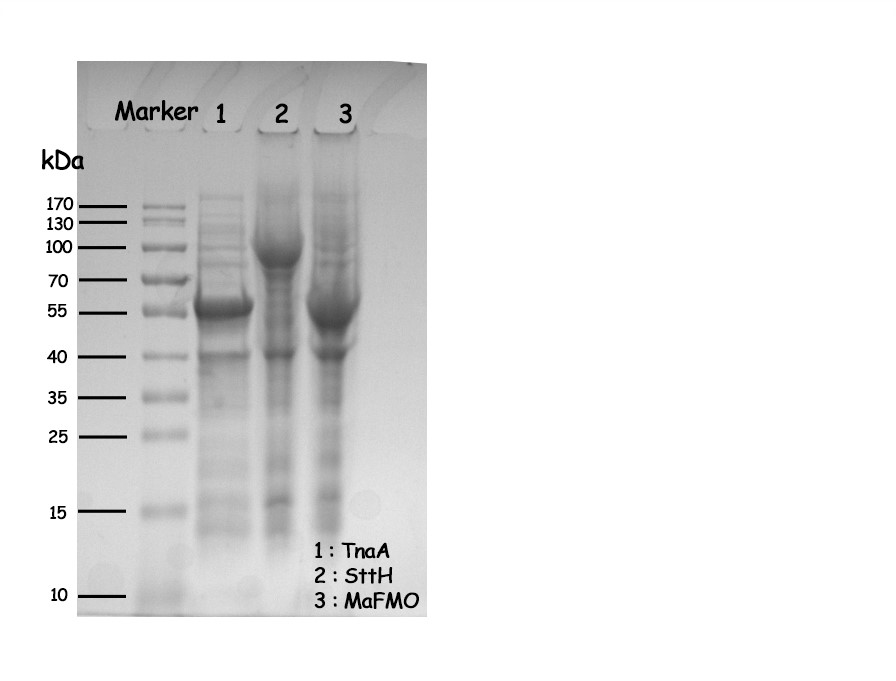
| + | <p>We used this part in our composite part BBa_K3722007, BBa_K3722008 and BBa_K3722009 to express tryptophanase (TnaA, BBa_K3722000), monooxygenase (MaFMO, BBa_K3722002) and tryptophan (Trp) halogenase (SttH, BBa_K3722003). All of them were constructed into pET-28a and transformed to E.coli (DE3) for expression.</p> | ||
| + | [[File:T--NWU-CHINA-A--SDS-PAGE result of TnaA, SttH, MaFMO.jpg|500px|center|]] | ||
| + | <p><center><b>Figure 1. SDS-PAGE result of TnaA, SttH, MaFMO. Lane on the far left is protein weight marker. Lane 1, tryptophanase (TnaA). Lane 2, tryptophan halogenase (SttH). Lane 3, monooxygenase (MaFMO).</b></center></p> | ||
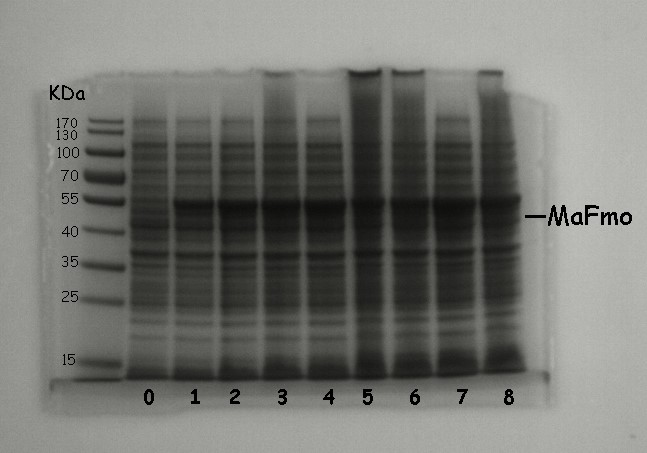
| + | [[File:T--NWU-CHINA-A--SDS-PAGE analysis of MaFMO.jpg|500px|center|]] | ||
| + | <p><center><b>Figure 2. SDS-PAGE analysis of MaFMO. Induction condition: LB media, OD600 0.6 ~ 0.8, IPTG 0.1 mM, 18°C and 200 rpm.</b></center></p> | ||
| + | <p><center><b>0:negative control 1:4h 2:5h 3:6h 4:7h 5:8h 6:9h 7:10h 8:11h | ||
| + | </b></center></p> | ||
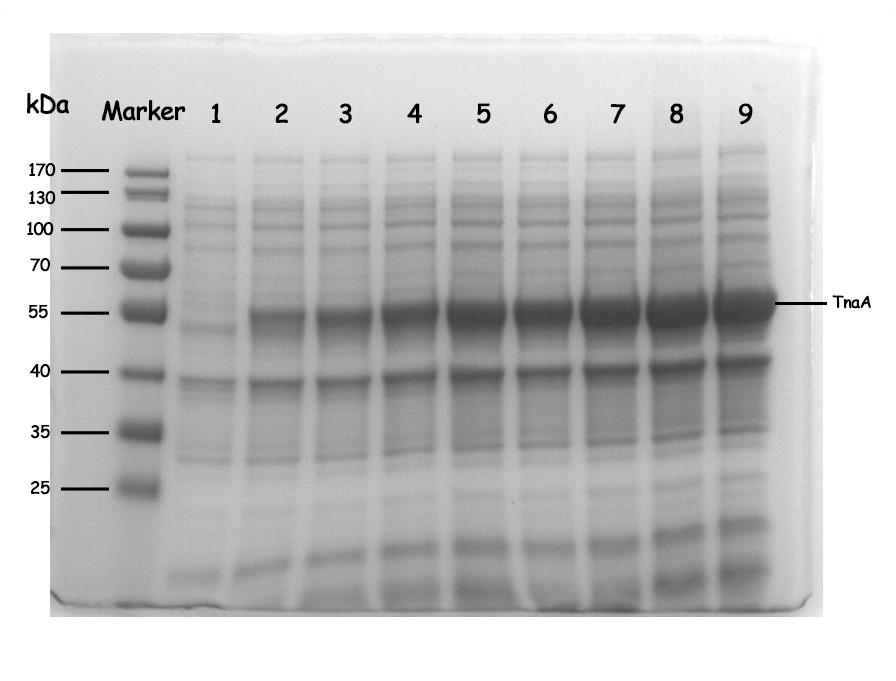
| + | [[File:T--NWU-CHINA-A--SDS-PAGE analysis of TnaA.jpg|500px|center|]] | ||
| + | <p><center><b>Figure 3. SDS-PAGE analysis of TnaA. Induction condition: LB media, OD600 0.6 ~ 0.8, IPTG 0.1 mM, 37 °C and 200 rpm.</b></center></p> | ||
| + | <p><center><b>1: negative control 2: 9h 3:10.5h 4:12h 5:13.5h 6:15h 7:16.5h 8: 18h 9: 19.5h | ||
| + | </b></center></p> | ||
| + | |||
| + | |||
| + | |||
| + | ===Characterization by NWU-CHINA-A 2022=== | ||
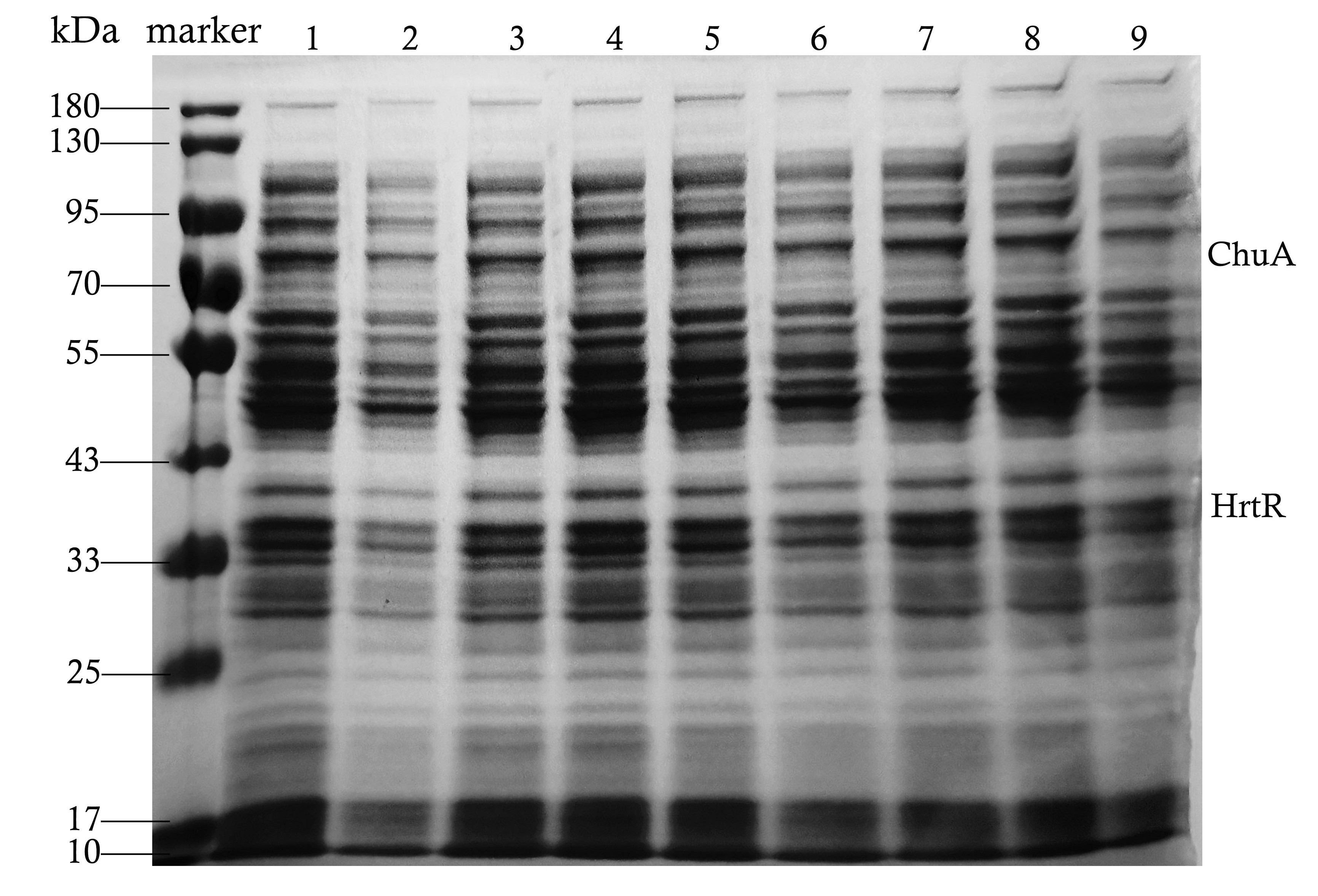
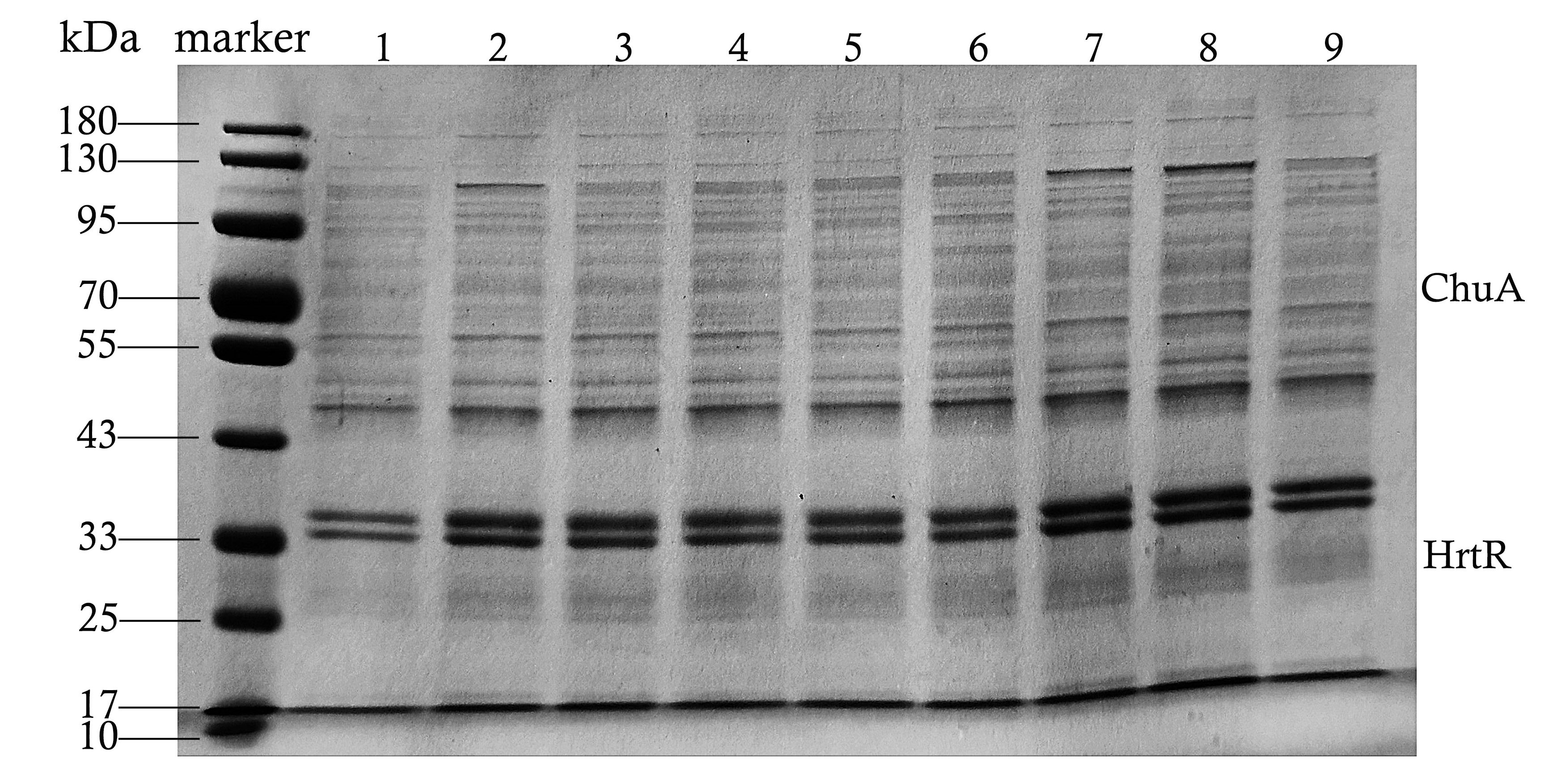
| + | <p>We used this part in our composite part BBa_K4199005, BBa_K4199006 and BBa_K4199007 to express ChuA(BBa_K4199001), HrtR(BBa_K4199002). All of them were constructed into pET-28a and transformed to E.coli BL21(DE3) for expression.</p> | ||
| + | [[File:NWU-CHINA-A--SDS-PAGE result of supernatant.jpg|500px|center|]] | ||
| + | <p><center><b>Figure 1. SDS-PAGE result of supernatant of bacteria broken by ultrasound.</b></center></p> | ||
| + | |||
| + | [[File:NWU-CHINA-A--SDS-PAGE result of precipitate.jpg|500px|center|]] | ||
| + | <p><center><b>Figure 2. SDS-PAGE results of resuspended precipitate after ultrasonic fragmentation of bacteria.</b></center></p> | ||
| + | |||
| + | |||
| − | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 08:49, 11 October 2022
A T7 promoter from pet28a plasmid.
Summary
It is a strong promoter from T7 bacteriophage that can specifically react to T7RNA polymerase. It is a sequence that initiates the transcription of T7 bacteriophage gene.
Characterization by NWU-CHINA-A 2021
We used this part in our composite part BBa_K3722007, BBa_K3722008 and BBa_K3722009 to express tryptophanase (TnaA, BBa_K3722000), monooxygenase (MaFMO, BBa_K3722002) and tryptophan (Trp) halogenase (SttH, BBa_K3722003). All of them were constructed into pET-28a and transformed to E.coli (DE3) for expression.
Characterization by NWU-CHINA-A 2022
We used this part in our composite part BBa_K4199005, BBa_K4199006 and BBa_K4199007 to express ChuA(BBa_K4199001), HrtR(BBa_K4199002). All of them were constructed into pET-28a and transformed to E.coli BL21(DE3) for expression.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]